MGMT promoter methylation in triple negative breast cancer of the GeparSixto trial
- PMID: 32841306
- PMCID: PMC7446962
- DOI: 10.1371/journal.pone.0238021
MGMT promoter methylation in triple negative breast cancer of the GeparSixto trial
Erratum in
-
Correction: MGMT promoter methylation in triple negative breast cancer of the GeparSixto trial.PLoS One. 2021 Sep 1;16(9):e0257142. doi: 10.1371/journal.pone.0257142. eCollection 2021. PLoS One. 2021. PMID: 34469495 Free PMC article.
Abstract
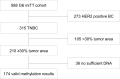
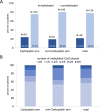
Triple-negative breast cancer (TNBC) is typically treated with chemotherapeutic agents, including carboplatin (Cb), an DNA platinating agent. The O6-methylguanine-DNA-methyltransferase gene (MGMT) encodes for the protein O6-alkylguanine-DNA-alkyltransferase (MGMT protein). MGMT protein is involved in DNA repair mechanisms to remove mutagenic and cytotoxic adducts from O6-guanine in DNA. In glioblastoma multiforme, MGMT methylation status is a predictive biomarker for increased response to temozolomide therapy. It has been suggested, that MGMT protein may have relevance for cellular adaptation and could have an influence on resistance to carboplatin therapy. We investigated the influence of MGMT promoter methylation on pathologic complete response and survival of patients with TNBC treated in the neoadjuvant GeparSixto trial. In 174 of 210 available TNBC tumors a valid MGMT promoter methylation status was determined by pyrosequencing of 5 CpG islands. In 21.8%, we detected a mean MGMT promoter methylation >10%. Overall, MGMT promoter methylation was not significantly associated with pathological complete response (pCR) rate. After stratification for the two therapy arms with and without Cb no statistically significant differences in therapy response rates between the two MGMT promoter methylation groups could be observed. Our results show that different MGMT promoter methylation status is not related to different chemotherapy response rates in the TNBC setting in GeparSixto.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: Dr. Schneeweiss reports grants from Celgene, grants from Roche, grants from AbbVie, grants from Molecular Partner, personal fees from Roche, personal fees from AstraZeneca, personal fees from Celgene, personal fees from Roche, personal fees from Roche, personal fees from Celgene, personal fees from Pfizer, personal fees from AstraZeneca, personal fees from Novartis, personal fees from MSD, personal fees from Tesaro, personal fees from Lilly, personal fees from Pfizer, other from Roche, outside the submitted work. Dr. v. Mackelenbergh reports travel grants and honoria from AstraZeneca, Amgen, Gebomic Health, Novartis, Lilly. Dr. Denkert reports personal fees from Novartis, personal fees from Roche, personal fees from MSD Oncology, from Daiichi Sankyo, grants from Myriad Genetics, other from Sividon Diagnostics / Myriad, outside the submitted work; In addition, Dr. Denkert has a patent EP18209672 pending, a patent EP20150702464 pending, and a patent Software (VMscope digital pathology) pending. All other authors declare no conflict of interest. We confirm that the patents do not alter our adherence to PLOS ONE policies on sharing data and materials. We would like to point out, that the patents had been added as general COI information but are not related to MGMT methylation.
Figures


Similar articles
-
Prevalence and clinicopathologic correlates of O⁶-methylguanine-DNA methyltransferase methylation status in patients with triple-negative breast cancer treated preoperatively by alkylating drugs.Clin Breast Cancer. 2014 Aug;14(4):285-90. doi: 10.1016/j.clbc.2014.02.010. Epub 2014 Mar 2. Clin Breast Cancer. 2014. PMID: 24709436
-
Methylation of O6-methylguanine-DNA methyltransferase (MGMT) promoter gene in triple-negative breast cancer patients.Breast Cancer Res Treat. 2012 Jul;134(1):131-7. doi: 10.1007/s10549-011-1945-9. Epub 2012 Jan 8. Breast Cancer Res Treat. 2012. PMID: 22228432
-
Detection of MGMT promoter methylation in glioblastoma using pyrosequencing.Int J Clin Exp Pathol. 2015 Feb 1;8(2):1790-6. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 25973069 Free PMC article.
-
The prognostic value of MGMT promoter methylation in glioblastoma: A meta-analysis of clinical trials.J Cell Physiol. 2018 Jan;233(1):378-386. doi: 10.1002/jcp.25896. Epub 2017 May 16. J Cell Physiol. 2018. PMID: 28266716 Review.
-
The prognostic value of MGMT promoter status by pyrosequencing assay for glioblastoma patients' survival: a meta-analysis.World J Surg Oncol. 2016 Oct 12;14(1):261. doi: 10.1186/s12957-016-1012-4. World J Surg Oncol. 2016. PMID: 27733166 Free PMC article. Review.
Cited by
-
Detection and relevance of epigenetic markers on ctDNA: recent advances and future outlook.Mol Oncol. 2021 Jun;15(6):1683-1700. doi: 10.1002/1878-0261.12978. Epub 2021 May 14. Mol Oncol. 2021. PMID: 33942482 Free PMC article. Review.
-
Underexplored reciprocity between genome-wide methylation status and long non-coding RNA expression reflected in breast cancer research: potential impacts for the disease management in the framework of 3P medicine.EPMA J. 2023 May 22;14(2):249-273. doi: 10.1007/s13167-023-00323-7. eCollection 2023 Jun. EPMA J. 2023. PMID: 37275549 Free PMC article. Review.
-
APC Promoter Methylation in Gastrointestinal Cancer.Front Oncol. 2021 Apr 23;11:653222. doi: 10.3389/fonc.2021.653222. eCollection 2021. Front Oncol. 2021. PMID: 33968756 Free PMC article. Review.
-
Tricarboxylic Acid Cycle Relationships with Non-Metabolic Processes: A Short Story with DNA Repair and Its Consequences on Cancer Therapy Resistance.Int J Mol Sci. 2024 Aug 21;25(16):9054. doi: 10.3390/ijms25169054. Int J Mol Sci. 2024. PMID: 39201738 Free PMC article. Review.
-
Correction: MGMT promoter methylation in triple negative breast cancer of the GeparSixto trial.PLoS One. 2021 Sep 1;16(9):e0257142. doi: 10.1371/journal.pone.0257142. eCollection 2021. PLoS One. 2021. PMID: 34469495 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

