Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents
- PMID: 32841542
- PMCID: PMC8601184
- DOI: 10.1111/1751-7915.13652
Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents
Abstract
DNA double-strand break (DSB) repair induced by the RNA-programmed nuclease Cas9 has become a popular method for genome editing. Direct genome editing via Cas9-CRISPR gRNA (guide RNA) ribonucleoprotein (RNP) complexes assembled in vitro has also been successful in some fungi. However, the efficiency of direct RNP transformation into fungal protoplasts is currently too low. Here, we report an optimized genome editing approach for filamentous fungi based on RNPs facilitated by adding chemical reagents. We increased the transformation efficiency of RNPs significantly by adding Triton X-100 and prolonging the incubation time, and the editing efficiency reached 100% in Trichoderma reesei and Cordyceps militaris. The optimized RNP-based method also achieved efficient (56.52%) homologous recombination integration with short homology arms (20 bp) and gene disruption (7.37%) that excludes any foreign DNA (selection marker) in T. reesei. In particular, after adding reagents related to mitosis and cell division, the further optimized protocol showed an increased ratio of edited homokaryotic transformants (from 0% to 40.0% for inositol and 71.43% for benomyl) from Aspergillus oryzae, which contains multinucleate spores and protoplasts. Furthermore, the multi-target engineering efficiency of the optimized RNP transformation method was similar to those of methods based on in vivo expression of Cas9. This newly established genome editing system based on RNPs may be widely applicable to construction of genome-edited fungi for the food and medical industries, and has good prospects for commercialization.
© 2020 The Authors. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

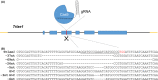

References
-
- Arazoe, T. , Miyoshi, K. , Yamato, T. , Ogawa, T. , Ohsato, S. , Arie, T. , and Kuwata, S. (2015) Tailor‐made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng 112: 2543–2549. - PubMed
-
- Arentshorst, M. , Ram, A.F. , and Meyer, V. (2012) Using non‐homologous end‐joining‐deficient strains for functional gene analyses in filamentous fungi. Methods Mol Biol 835: 133–150. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

