Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae
- PMID: 32842463
- PMCID: PMC7558325
- DOI: 10.3390/jof6030144
Phylogenetic and Chemotaxonomic Studies Confirm the Affinities of Stromatoneurospora phoenix to the Coprophilous Xylariaceae
Abstract
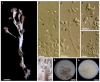
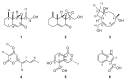
The genus Stromatoneurospora was erected in 1973 by Jong and Davis to accommodate the pyrophilic pyrenomycete Sphaeria phoenix and has traditionally been placed in the family Xylariaceae based on morphological features. However, no living culture of this genus has so far been available in the public domain. Molecular data were restricted to an internal transcribed spacer (ITS) sequence that only confirmed the familial position, and was generated from a strain that is not deposited in a public culture collection. We have recently collected fresh material and were able to culture this fungus from Thailand. The secondary metabolites of this strains were analysed after fermentation in multiple media. The the prominent components of these fermentation were purified, using preparative chromatography. Aside from two new eremophilane sesquiterpenoids named phoenixilanes A-B (1-2), four other components that are known from species of the xylariaceous genera Xylaria and Poronia were identified by spectral methods (nuclear magnetic resonance spectroscopy and high resolution mass spectrometry). Notably, (-)-(R)-6-hydroxy-3-methyl-4-dihydroisocoumarin-5-carboxylic acid (6) has not been reported as a natural product before. Moreover, DNA sequences of Stromatoneurospora phoenix clustered with members of the genera Poronia and Podosordaria in a multi-locus molecular phylogeny. These results confirmed that the genus belongs to the same evolutionary lineage as the coprophilic Xylariaceae. The results also suggest that this lineage has evolved independently from the plant-inhabiting saprotrophs and endophytes that are closely related to the genus Xylaria. These findings are discussed in relation to some theories about the endophytic vs. the pyrophilic/coprophilic fungal life style.
Keywords: Sordariomycetes; Xylariales; secondary metabolites; sesquiterpenoids; structure elucidation.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
-
- Jong S.C., Davis E.E. Stromatic Neurosporas. Mycologia. 1973;65:458–464. doi: 10.1080/00275514.1973.12019453. - DOI
-
- Fries E.M. Eclogae fungorum, praecipue ex herbarus germanorum de scriptorum. Linnaea. 1830;5:497–553.
-
- Daranagama D.A., Hyde K.D., Sir E.B., Thambugala K.M., Tian Q., Samarakoon M.C., McKenzie E.H.C., Jayasiri S.C., Tibpromma S., Bhat J.D., et al. Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Divers. 2017;88:1–165. doi: 10.1007/s13225-017-0388-y. - DOI
-
- Wendt L., Sir E.B., Kuhnert E., Heitkämper S., Lambert C., Hladki A.I., Romero A.I., Luangsa-ard J.J., Srikitikulchai P., Peršoh D., et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018;17:115–154. doi: 10.1007/s11557-017-1311-3. - DOI
-
- Peláez F., González V., Platas G., Sánchez Ballesteros J., Rubio V. Molecular phylogenetic studies within the Xylariaceae based on ribosomal DNA sequences. Fungal Divers. 2008;31:111–134.
Grants and funding
LinkOut - more resources
Full Text Sources

