Defining Caenorhabditis elegans as a model system to investigate lipoic acid metabolism
- PMID: 32843480
- PMCID: PMC7606682
- DOI: 10.1074/jbc.RA120.013760
Defining Caenorhabditis elegans as a model system to investigate lipoic acid metabolism
Abstract
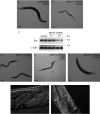
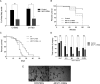
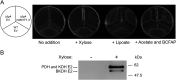
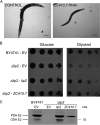
Lipoic acid (LA) is a sulfur-containing cofactor that covalently binds to a variety of cognate enzymes that are essential for redox reactions in all three domains of life. Inherited mutations in the enzymes that make LA, namely lipoyl synthase, octanoyltransferase, and amidotransferase, result in devastating human metabolic disorders. Unfortunately, because many aspects of this essential pathway are still obscure, available treatments only serve to alleviate symptoms. We envisioned that the development of an organismal model system might provide new opportunities to interrogate LA biochemistry, biology, and physiology. Here we report our investigations on three Caenorhabditis elegans orthologous proteins involved in this post-translational modification. We established that M01F1.3 is a lipoyl synthase, ZC410.7 an octanoyltransferase, and C45G3.3 an amidotransferase. Worms subjected to RNAi against M01F1.3 and ZC410.7 manifest larval arrest in the second generation. The arrest was not rescued by LA supplementation, indicating that endogenous synthesis of LA is essential for C. elegans development. Expression of the enzymes M01F1.3, ZC410.7, and C45G3.3 completely rescue bacterial or yeast mutants affected in different steps of the lipoylation pathway, indicating functional overlap. Thus, we demonstrate that, similarly to humans, C. elegans is able to synthesize LA de novo via a lipoyl-relay pathway, and suggest that this nematode could be a valuable model to dissect the role of protein mislipoylation and to develop new therapies.
Keywords: Caenorhabditis elegans (C. elegans); RNA interference; RNA interference (RNAi); development; energy metabolism; fatty acid metabolism; inborn error of metabolism; lipoic acid; mitochondrial metabolism; oxidative stress; post-translational modification.
© 2020 Lavatelli et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Comment in
-
Shining light on rhodopsin selectivity: How do proteins decide whether to transport H+ or Cl-?J Biol Chem. 2020 Oct 30;295(44):14805-14806. doi: 10.1074/jbc.H120.016032. J Biol Chem. 2020. PMID: 33127867 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

