Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLpro Reporter Assay
- PMID: 32843534
- PMCID: PMC7592234
- DOI: 10.1128/JVI.01265-20
Development of a Fluorescence-Based, High-Throughput SARS-CoV-2 3CLpro Reporter Assay
Abstract
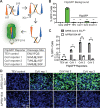
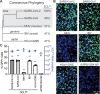
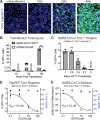
In late 2019, a human coronavirus, now known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged, likely from a zoonotic reservoir. This virus causes COVID-19, has infected millions of people, and has led to hundreds of thousands of deaths across the globe. While the best interventions to control and ultimately stop the pandemic are prophylactic vaccines, antiviral therapeutics are important to limit morbidity and mortality in those already infected. At this time, only one FDA-approved anti-SARS-CoV-2 antiviral drug, remdesivir, is available, and unfortunately, its efficacy appears to be limited. Thus, the identification of new and efficacious antivirals is of the highest importance. In order to facilitate rapid drug discovery, flexible, sensitive, and high-throughput screening methods are required. With respect to drug targets, most attention is focused on either the viral RNA-dependent RNA polymerase or the main viral protease, 3CLpro 3CLpro is an attractive target for antiviral therapeutics, as it is essential for processing newly translated viral proteins and the viral life cycle cannot be completed without protease activity. In this work, we report a new assay to identify inhibitors of 3CLpro Our reporter is based on a green fluorescent protein (GFP)-derived protein that fluoresces only after cleavage by 3CLpro This experimentally optimized reporter assay allows for antiviral drug screening in human cell culture at biosafety level 2 (BSL2) with high-throughput compatible protocols. Using this screening approach in combination with existing drug libraries may lead to the rapid identification of novel antivirals to suppress SARS-CoV-2 replication and spread.IMPORTANCE The COVID-19 pandemic has already led to more than 700,000 deaths and innumerable changes to daily life worldwide. Along with development of a vaccine, identification of effective antivirals to treat infected patients is of the highest importance. However, rapid drug discovery requires efficient methods to identify novel compounds that can inhibit the virus. In this work, we present a method for identifying inhibitors of the SARS-CoV-2 main protease, 3CLpro This reporter-based assay allows for antiviral drug screening in human cell culture at biosafety level 2 (BSL2) with high-throughput compatible sample processing and analysis. This assay may help identify novel antivirals to control the COVID-19 pandemic.
Keywords: FlipGFP; antivirals; coronavirus; protease; screening.
Copyright © 2020 American Society for Microbiology.
Figures
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. doi:10.1056/NEJMoa2001316. - DOI - PMC - PubMed
-
- Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S, Tsoi H-W, Lo SK-F, Chan K-H, Poon VK-M, Chan W-M, Ip JD, Cai J-P, Cheng VC-C, Chen H, Hui CK-M, Yuen K-Y. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. doi:10.1016/S0140-6736(20)30154-9. - DOI - PMC - PubMed
-
- Johns Hopkins Coronavirus Resource Center. 2020. COVID-19 map. https://coronavirus.jhu.edu/map.html.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

