Puromycin reactivity does not accurately localize translation at the subcellular level
- PMID: 32844748
- PMCID: PMC7490009
- DOI: 10.7554/eLife.60303
Puromycin reactivity does not accurately localize translation at the subcellular level
Abstract
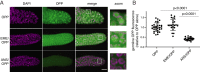
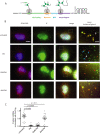
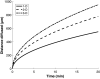
Puromycin is a tyrosyl-tRNA mimic that blocks translation by labeling and releasing elongating polypeptide chains from translating ribosomes. Puromycin has been used in molecular biology research for decades as a translation inhibitor. The development of puromycin antibodies and derivatized puromycin analogs has enabled the quantification of active translation in bulk and single-cell assays. More recently, in vivo puromycylation assays have become popular tools for localizing translating ribosomes in cells. These assays often use elongation inhibitors to purportedly inhibit the release of puromycin-labeled nascent peptides from ribosomes. Using in vitro and in vivo experiments in various eukaryotic systems, we demonstrate that, even in the presence of elongation inhibitors, puromycylated peptides are released and diffuse away from ribosomes. Puromycylation assays reveal subcellular sites, such as nuclei, where puromycylated peptides accumulate post-release and which do not necessarily coincide with sites of active translation. Our findings urge caution when interpreting puromycylation assays in vivo.
Keywords: C. elegans; biochemistry; cell biology; chemical biology; emetine; human; op-puro; puromycin; puromycylation; ribosome.
© 2020, Enam et al.
Conflict of interest statement
SE, BZ, DG, MC, NL, GS No competing interests declared, RG Reviewing editor, eLife
Figures
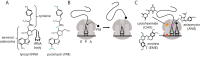


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

