Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection
- PMID: 32845525
- PMCID: PMC8078202
- DOI: 10.1002/14651858.CD013705
Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection
Update in
-
Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2021 Mar 24;3(3):CD013705. doi: 10.1002/14651858.CD013705.pub2. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 Jul 22;7:CD013705. doi: 10.1002/14651858.CD013705.pub3. PMID: 33760236 Free PMC article. Updated.
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the resulting COVID-19 pandemic present important diagnostic challenges. Several diagnostic strategies are available to identify or rule out current infection, identify people in need of care escalation, or to test for past infection and immune response. Point-of-care antigen and molecular tests to detect current SARS-CoV-2 infection have the potential to allow earlier detection and isolation of confirmed cases compared to laboratory-based diagnostic methods, with the aim of reducing household and community transmission.
Objectives: To assess the diagnostic accuracy of point-of-care antigen and molecular-based tests to determine if a person presenting in the community or in primary or secondary care has current SARS-CoV-2 infection.
Search methods: On 25 May 2020 we undertook electronic searches in the Cochrane COVID-19 Study Register and the COVID-19 Living Evidence Database from the University of Bern, which is updated daily with published articles from PubMed and Embase and with preprints from medRxiv and bioRxiv. In addition, we checked repositories of COVID-19 publications. We did not apply any language restrictions.
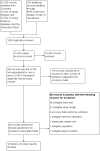
Selection criteria: We included studies of people with suspected current SARS-CoV-2 infection, known to have, or not to have SARS-CoV-2 infection, or where tests were used to screen for infection. We included test accuracy studies of any design that evaluated antigen or molecular tests suitable for a point-of-care setting (minimal equipment, sample preparation, and biosafety requirements, with results available within two hours of sample collection). We included all reference standards to define the presence or absence of SARS-CoV-2 (including reverse transcription polymerase chain reaction (RT-PCR) tests and established clinical diagnostic criteria).
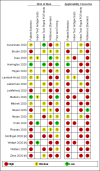
Data collection and analysis: Two review authors independently screened studies and resolved any disagreements by discussion with a third review author. One review author independently extracted study characteristics, which were checked by a second review author. Two review authors independently extracted 2x2 contingency table data and assessed risk of bias and applicability of the studies using the QUADAS-2 tool. We present sensitivity and specificity, with 95% confidence intervals (CIs), for each test using paired forest plots. We pooled data using the bivariate hierarchical model separately for antigen and molecular-based tests, with simplifications when few studies were available. We tabulated available data by test manufacturer.
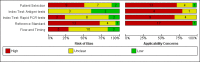
Main results: We included 22 publications reporting on a total of 18 study cohorts with 3198 unique samples, of which 1775 had confirmed SARS-CoV-2 infection. Ten studies took place in North America, two in South America, four in Europe, one in China and one was conducted internationally. We identified data for eight commercial tests (four antigen and four molecular) and one in-house antigen test. Five of the studies included were only available as preprints. We did not find any studies at low risk of bias for all quality domains and had concerns about applicability of results across all studies. We judged patient selection to be at high risk of bias in 50% of the studies because of deliberate over-sampling of samples with confirmed COVID-19 infection and unclear in seven out of 18 studies because of poor reporting. Sixteen (89%) studies used only a single, negative RT-PCR to confirm the absence of COVID-19 infection, risking missing infection. There was a lack of information on blinding of index test (n = 11), and around participant exclusions from analyses (n = 10). We did not observe differences in methodological quality between antigen and molecular test evaluations. Antigen tests Sensitivity varied considerably across studies (from 0% to 94%): the average sensitivity was 56.2% (95% CI 29.5 to 79.8%) and average specificity was 99.5% (95% CI 98.1% to 99.9%; based on 8 evaluations in 5 studies on 943 samples). Data for individual antigen tests were limited with no more than two studies for any test. Rapid molecular assays Sensitivity showed less variation compared to antigen tests (from 68% to 100%), average sensitivity was 95.2% (95% CI 86.7% to 98.3%) and specificity 98.9% (95% CI 97.3% to 99.5%) based on 13 evaluations in 11 studies of on 2255 samples. Predicted values based on a hypothetical cohort of 1000 people with suspected COVID-19 infection (with a prevalence of 10%) result in 105 positive test results including 10 false positives (positive predictive value 90%), and 895 negative results including 5 false negatives (negative predictive value 99%). Individual tests We calculated pooled results of individual tests for ID NOW (Abbott Laboratories) (5 evaluations) and Xpert Xpress (Cepheid Inc) (6 evaluations). Summary sensitivity for the Xpert Xpress assay (99.4%, 95% CI 98.0% to 99.8%) was 22.6 (95% CI 18.8 to 26.3) percentage points higher than that of ID NOW (76.8%, (95% CI 72.9% to 80.3%), whilst the specificity of Xpert Xpress (96.8%, 95% CI 90.6% to 99.0%) was marginally lower than ID NOW (99.6%, 95% CI 98.4% to 99.9%; a difference of -2.8% (95% CI -6.4 to 0.8)) AUTHORS' CONCLUSIONS: This review identifies early-stage evaluations of point-of-care tests for detecting SARS-CoV-2 infection, largely based on remnant laboratory samples. The findings currently have limited applicability, as we are uncertain whether tests will perform in the same way in clinical practice, and according to symptoms of COVID-19, duration of symptoms, or in asymptomatic people. Rapid tests have the potential to be used to inform triage of RT-PCR use, allowing earlier detection of those testing positive, but the evidence currently is not strong enough to determine how useful they are in clinical practice. Prospective and comparative evaluations of rapid tests for COVID-19 infection in clinically relevant settings are urgently needed. Studies should recruit consecutive series of eligible participants, including both those presenting for testing due to symptoms and asymptomatic people who may have come into contact with confirmed cases. Studies should clearly describe symptomatic status and document time from symptom onset or time since exposure. Point-of-care tests must be conducted on samples according to manufacturer instructions for use and be conducted at the point of care. Any future research study report should conform to the Standards for Reporting of Diagnostic Accuracy (STARD) guideline.
Copyright © 2020 The Authors. Cochrane Database of Systematic Reviews published by John Wiley & Sons, Ltd. on behalf of The Cochrane Collaboration.
Conflict of interest statement
Jonathan J Deeks: none known
Jacqueline Dinnes: none known
Yemisi Takwoingi: none known
Clare Davenport: none known
Mariska MG Leeflang: none known
René Spijker: the Dutch Cochrane Centre (DCC) has received grants for performing commissioned systematic reviews. In no situation, the commissioner had any influence on the results of the work.
Lotty Hooft: none known
Ann Van den Bruel: none known
Devy Emperador: is employed by FIND with funding from DFID and KFW. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high quality diagnostic tools for low resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. .FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Sabine Dittrich: is employed by FIND with funding from DFID and Australian Aid. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high quality diagnostic tools for low resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. .FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Ada Adriano: none known
Sophie Beese: none known
Janine Dretzke: none known
Lavinia Ferrante di Ruffano: none known
Isobel Harris: none known
Malcolm Price: none known
Sian Taylor‐Phillips: none known
Sarah Berhane: is funded by NIHR Birmingham Biomedical Research Centre.
Jane Cunningham: none known
Figures
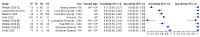








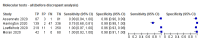
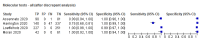
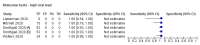
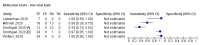
Comment in
-
Rapid antigen and molecular tests had varied sensitivity and ≥97% specificity for detecting SARS-CoV-2 infection.Ann Intern Med. 2020 Dec 15;173(12):JC69. doi: 10.7326/ACPJ202012150-069. Ann Intern Med. 2020. PMID: 33316181
References
References to studies included in this review
Assennato 2020 {published data only}
Broder 2020 {published data only}
Diao 2020 {published data only}
-
- Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv [Preprint] 10 March 2020:1-13. [DOI: 10.1101/2020.03.07.20032524] - DOI
Harrington 2020 {published data only}
-
- Harrington A, Cox B, Snowdon J, Bakst J, Ley E, Grajales P, et al. Comparison of Abbott ID NOW and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. Journal of Clinical Microbiology 2020;58(8):e00798-20. [DOI: 10.1128/JCM.00798-20.] - DOI - PMC - PubMed
Hogan 2020 {published data only}
Lambert‐Niclot 2020 {published data only}
-
- Lambert-Niclot S, Cuffel A, Le Pape S, Vauloup-Fellous C, Morand-Joubert L, Roque-Afonso AM, et al. Evaluation of a rapid diagnostic assay for detection of SARS CoV-2 antigen in nasopharyngeal swab. Journal of Clinical Microbiology 2020;58(8):e00977-20. [DOI: 10.1128/JCM.00977-20] - DOI - PMC - PubMed
Lieberman 2020 {published data only}
-
- Lieberman JA, Pepper G, Naccache SN, Huang ML, Jerome KR, Greninger AL. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. Journal of Clinical Microbiology 2020;58(8):e00821-20. [DOI: 10.1128/JCM.00821-20] - DOI - PMC - PubMed
Loeffelholz 2020 {published data only}
Mertens 2020 {published data only}
Mitchell 2020 {published data only}
Moore 2020 {published data only}
-
- Moore NM, Li H, Schejbal D, Lindsley J, Hayden M. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCOV RT-PCR assay for the qualitative detection of SARS-CoV-2 from upper respiratory tract specimens. medRxiv [Preprint] 2020:1-22. [DOI: 10.1101/2020.05.02.20088740] - DOI - PMC - PubMed
Moran 2020 {published data only}
Porte 2020 {published data only}
-
- Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, et al. Evaluation of novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. papers.ssrn.com/abstract=3569871 14 April 2020:1-23. - PMC - PubMed
Rhoads 2020 {published data only}
-
- Rhoads DD, Cherian SS, Roman K, Stempak LM, Schmotzer CL, Sadri N. Comparison of Abbott ID NOW, Diasorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. Journal of Clinical Microbiology 2020;58(8):e00760-20. [DOI: 10.1128/JCM.00760-20] - DOI - PMC - PubMed
Smithgall 2020 [A] {published data only}
Smithgall 2020 [B] {published data only}
Weitzel 2020 [A] {published data only}
Weitzel 2020 [B] {published data only}
Weitzel 2020 [C] {published data only}
Weitzel 2020 [D] {published data only}
Wolters 2020 {published data only}
-
- Wolters F, Van de Bovenkamp J, Van den Bosch B, Van den Brink S, Broeders M, Chung NH, et al. Multi-center evaluation of Cepheid Xpert(R) Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. Journal of Clinical Virology 2020;128:104426. [DOI: 10.1016/j.jcv.2020.104426] - DOI - PMC - PubMed
Zhen 2020 [A] {published data only}
References to studies excluded from this review
Ai 2020 {published data only}
-
- Ai JW, Zhang HC, Xu T, Wu J, Zhu M, Yu YQ, et al. Optimizing diagnostic strategy for novel coronavirus pneumonia, a multi-center study in Eastern China. medRxiv [Preprint] 17 February 2020:1-18. [DOI: 10.1101/2020.02.13.20022673] - DOI
Anahtar 2020 {published data only}
-
- Anahtar MN, McGrath GE, Rabe BA, Tanner NA, White BA, Lennerz JK, et al. Clinical assessment and validation of a rapid and sensitive SARS-CoV-2 test using reverse-transcription loop-mediated isothermal amplification. medRxiv [Preprint] 18 May 2020:1-22. [DOI: 10.1101/2020.05.12.20095638] - DOI - PMC - PubMed
Arumugam 2020 {published data only}
Baek 2020 {published data only}
Barra 2020 {published data only}
Basu 2020 {published data only}
-
- Basu A, Zinger T, Inglima K, Woo KM, Atie O, Yurasits L, et al. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test in nasopharyngeal swabs transported in viral media and dry nasal swabs, in a New York City academic institution. Journal of Clinical Microbiology 2020;58(8):e01136-20. [DOI: 10.1128/JCM.01136-20] - DOI - PMC - PubMed
Behrmann 2020 {published data only}
-
- Behrmann O, Bachmann I, Spiegel M, Schramm M, El Wahed AA, Dobler G, et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ). Clinical Chemistry 8 May 2020 [Epub ahead of print]:hvaa116. [DOI: 10.1093/clinchem/hvaa116] - DOI - PMC - PubMed
Bordi 2020 {published data only}
Broughton 2020 {published data only}
-
- Broughton JP, Deng X, Yu G, Fasching CL, Singh J, Streithorst J, et al. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. medRxiv [Preprint] 27 March 2020:1-28. [DOI: 10.1101/2020.03.06.20032334] - DOI
Callahan 2020 {published data only}
-
- Callahan CJ, Lee R, Zulauf K, Tamburello L, Smith KP, Previtera J, et al. Open development and clinical validation of multiple 3D-printed sample-collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. medRxiv [Preprint] 7 May 2020:1-16. [EMBASE: 10.1101/2020.04.14.20065094] - PMC - PubMed
-
- Callahan CJ, Lee R, Zulauf KE, Tamburello L, Smith KP, Previtera J, et al. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. Journal of Clinical Microbiology 2020;58(8):e00876-20. [DOI: 10.1128/JCM.00876-20] - DOI - PMC - PubMed
Chandler‐Brown 2020 {published data only}
-
- Chandler-Brown D, Bueno AM, Atay O, Tsao DS. A highly scalable and rapidly deployable RNA extraction-free COVID-19 assay by quantitative Sanger sequencing. medRxiv [Preprint] 10 April 2020:1-15. [DOI: 10.1101/2020.04.07.029199] - DOI
Colson 2020 {published data only}
Comar 2020 {published data only}
-
- Comar M, Brumat M, Concas MP, Argentini G, Bianco A, Bicego L, et al. COVID-19 experience: first Italian survey on healthcare staff members from a Mother-Child Research hospital using combined molecular and rapid immunoassays test. medRxiv [Preprint] 22 April 2020:1-12. [DOI: 10.1101/2020.04.19.20071563] - DOI
Crone 2020 {published data only}
-
- Crone MA, Priestman M, Ciechonska M, Jensen K, Sharp DJ, Randell P, et al. A new role for Biofoundries in rapid prototyping, development, and validation of automated clinical diagnostic tests for SARS-CoV-2. medRxiv [Preprint] 12 May 2020:1-31. [DOI: 10.1101/2020.05.02.20088344] - DOI
Curti 2020 {published data only}
-
- Curti L, Pereyra-Bonnet F, Gimenez CA. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv [Preprint] 2 March 2020:1-10. [DOI: 10.1101/2020.02.29.971127] - DOI
Ding 2020 {published data only}
-
- Ding X, Yin K, Li Z, Liu C. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv [Preprint] 21 March 2020:1-19. [DOI: 10.1101/2020.03.19.998724] - DOI
Dohla 2020 {published data only}
Farfan 2020 {published data only}
Francis 2020 {published data only}
-
- Francis R, Le Bideau M, Jardot P, Grimaldier C, Raoult D, Khalil JY, et al. High speed large scale automated isolation of SARS-CoV-2 from clinical samples using miniaturized co-culture coupled with high content screening. bioRxiv [Preprint] 19 May 2020:1-23. [DOI: 10.1101/2020.05.14.097295] - DOI - PMC - PubMed
Freire‐Paspuel 2020 {published data only}
-
- Freire-Paspuel B, Vega-Marino P, Velez A, Cruz M, Bereguiain MA. High sensitivity CDC EUA SARS-CoV-2 kit-based End Point-PCR assay. medRxiv [Preprint] 18 May 2020:1-7. [DOI: 10.1101/2020.05.11.20098590] - DOI
Ganguli 2020 {published data only}
Giamarellos‐Bourboulis 2020 {published data only}
Gonzalez‐Gonzalez 2020 {published data only}
-
- Gonzalez-Gonzalez E, Lara-Mayorga IM, Rodriguez-Sanchez IP, Yee-de Leon F, Garcia-Rubio A, Garciamendez-Mijares CE, et al. Scaling diagnostics in times of COVID-19: rapid prototyping of 3D-printed water circulators for Loop-mediated Isothermal Amplification (LAMP) and detection of SARS-CoV-2 virus. medRxiv [Preprint] 19 June 2020:1-39. [DOI: 10.1101/2020.04.09.20058651] - DOI
Grant 2020 {published data only}
-
- Grant PR, Turner MA, Shin GY, Nastouli E, Levett LJ. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. bioRxiv [Preprint] 9 April 2020:1-6.
Hass 2020 {published data only}
Hogan 2020a {published data only}
Hu 2020 {published data only}
Huang 2020 {published data only}
Jiang 2020 {published data only}
-
- Jiang M, Pan W, Arastehfar A, Fang W, ling L, Fang H, et al. Development and validation of a rapid single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. medRxiv [Preprint] 27 March 2020:1-12. [DOI: 10.1101/2020.03.15.20036376] - DOI - PMC - PubMed
Joung 2020 {published data only}
-
- Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang MW, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv [Preprint] 8 May 2020:1-21. [DOI: 10.1101/2020.05.04.20091231] - DOI
Kalikiri 2020 {published data only}
-
- Kalikiri MK, Hasan M, Mirza F, Xaba T, Tang P, Lorenz S. High-throughput extraction of SARS-CoV-2 RNA from nasopharyngeal swabs using solid-phase reverse immobilization beads. medRxiv [Preprint] 11 April 2020:1-5. [DOI: 10.1101/2020.04.08.20055731] - DOI
Kim 2019 {published data only}
Konrad 2020 {published data only}
Kurstjens 2020 {published data only}
Lalli 2020 {published data only}
Lamb 2020 {published data only}
Lee 2020 {published data only}
-
- Lee JY, Best N, McAuley J, Porter JL, Seemann T, Schultz MB, et al. Validation of a single-step, single-tube reverse transcription-loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. bioRxiv [Preprint] 30 April 2020:1-32. [DOI: 10.1101/2020.04.28.067363] - DOI - PMC - PubMed
Lin 2020 {published data only}
Lowe 2020 {published data only}
Lu 2020 {published data only}
Lu 2020a {published data only}
Mahari 2020 {published data only}
-
- Mahari S, Roberts A, Shahdeo D, Gandhi S. eCovSens-Ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCOVID-19 antigen, a spike protein domain 1 of SARS-CoV-2. bioRxiv [Preprint] 11 May 2020:1-20. [DOI: 10.1101/2020.04.24.059204] - DOI
Marzinotto 2020 {published data only}
McCormick‐Baw 2020 {published data only}
-
- McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, et al. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. Journal of Clinical Microbiology 2020;58(8):e01109-20. [DOI: 10.1128/JCM.01109-20] - DOI - PMC - PubMed
McRae 2020 {published data only}
Mei 2020 {published data only}
Noerz 2020 {published data only}
Osterdahl 2020 {published data only}
Paden 2020 {published data only}
Pellanda 2020 {published data only}
-
- Pellanda LC, Wendland EM, McBride AJ, Tovo-Rodrigues L, Ferreira MR, Dellagostin OA, et al. Sensitivity and specificity of a rapid test for assessment of exposure to SARS-CoV-2 in a community-based setting in Brazil. medRxiv [Preprint] 10 May 2020:1-10. [DOI: 10.1101/2020.05.06.20093476] - DOI
Pfefferle 2020 {published data only}
Seo 2020 {published data only}
-
- Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020;14(4):5135-42. - PubMed
Smyrlaki 2020 {published data only}
St Hilaire 2020 {published data only}
Tan 2020 {published data only}
-
- Tan X, Lin C, Zhang J, Khaing OM, Fan X. Rapid and quantitative detection of COVID-19 markers in micro-liter sized samples. bioRxiv [Preprint] 22 April 2020:1-17. [DOI: 10.1101/2020.04.20.052233] - DOI
Visseaux 2020 {published data only}
-
- Visseaux B, Le Hingrat Q, Collin G, Bouzid D, Lebourgeois S, Le Pluart D, et al. Evaluation of the QIAstat-Dx Respiratory SARS-CoV-2 Panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. Journal of Clinical Microbiology 2020;58(8):e00630-20. [DOI: 10.1128/JCM.00630-20] - DOI - PMC - PubMed
Wang 2020 {published data only}
Wang 2020a {published data only}
Wee 2020 {published data only}
Xue 2020 {published data only}
Yan 2020 {published data only}
Yang 2020 {published data only}
-
- Yang W, Dang X, Wang Q, Xu M, Zhao Q, Zhou Y, et al. Rapid detection of SARS-CoV-2 using reverse transcription RT-LAMP method. medRxiv [Preprint] 3 March 2020:1-25. [DOI: 10.1101/2020.03.02.20030130] - DOI
Yu 2020 {published data only}
Yu 2020a {published data only}
-
- Yu L, Wu S, Hao X, Li X, Liu X, Ye S, et al. Rapid colorimetric detection of COVID-19 coronavirus using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) diagnostic platform: iLACO. medRxiv [Preprint] 24 February 2020:1-19. [DOI: 10.1101/2020.02.20.20025874] - DOI - PMC - PubMed
Zamecnik 2020 {published data only}
Zeng 2020 {published data only}
Zhang 2020 {published data only}
-
- Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba RO, Wei H, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv [Preprint] 29 February 2020:1-14. [DOI: 10.1101/2020.02.26.20028373] - DOI
Zhao 2020 {published data only}
-
- Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv [Preprint] 27 February 2020:1-18. [DOI: 10.1101/2020.02.22.961268] - DOI
Additional references
Arevalo‐Rodriguez 2020
Bossuyt 2015
Carter 2020
Cheng 2020
Corman 2020
-
- Corman V, Bleicker T, Brünink S, Drosten C, Landt O, Koopmans M, et al. Diagnostic detection of Wuhan coronavirus 2019 by real-time RT-PCR - protocol and preliminary evaluation as of Jan 13, 2020. Available from www.who.int/docs/default-source/coronaviruse/wuhan-virus-assay-v1991527e... 2020.
Covidence [Computer program]
-
- Veritas Health Innovation Covidence. Version accessed 27 April 2020. Melbourne, Australia: Veritas Health Innovation. Available at covidence.org.
Deeks 2020
-
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. Art. No: CD013596. [DOI: 10.1002/14651858.CD013596] - DOI
Deeks 2020a
FIND 2020
-
- FIND. SARS-COV-2 Diagnostic pipeline. www.finddx.org/covid-19/pipeline/ Accessed 19 July 2020.
Green 2020
-
- Green K, Graziadio S, Turner P, Fanshawe T, Allen J, on behalf of the Oxford COVID-19 Evidence Service Team Centre. Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Available at www.cebm.net/oxford-covid-19/ 7 April 2020.
Hadgu 1999
Kozel 2017
Leeflang 2013
McInnes 2018
McInnes 2020
Moher 2009
Pai 2012
Reitsma 2005
Review Manager 2014 [Computer program]
-
- Nordic Cochrane Centre, The Cochrane Collaboration Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Stata [Computer program]
-
- Stata. Version 15. College Station, TX, USA: StataCorp, 2017. Available at www.stata.com.
Struyf 2020
-
- Struyf T, Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19 disease. Cochrane Database of Systematic Reviews 2020, Issue 7. Art. No: CD013665. [DOI: 10.1002/14651858.CD013665] - DOI - PMC - PubMed
Subsoontorn 2020
Takwoingi 2017
Usher‐Smith 2016
Whiting 2011
-
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. - PubMed
WHO 2018
-
- World Health Organization (WHO). Diagnostic assessment: in vitro diagnostic medical devices (IVDs) used for the detection of high-risk human papillomavirus (HPV) genotypes in cervical cancer screening. Licence: CC BY-NC-SA 3.0 IGO. Available at apps.who.int/iris/handle/10665/272282 2018.
WHO 2020a
-
- World Health Organization. Laboratory testing of 2019 novel coronavirus (2019-nCoV) in suspected human cases: interim guidance. Available from www.who.int/publications/i/item/laboratory-testing-for-2019-novel-corona... 2020.
WHO 2020b
-
- Global surveillance for COVID-19 caused by human infection with COVID-19 virus: interim guidance. Available from www.who.int/publications/i/item/global-surveillance-for-covid-19-caused-... 2020.
WHO 2020c
-
- COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.0.1. Available from www.who.int/publications/m/item/covid-19-target-product-profiles-for-pri... 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

