Dynamics of Ionic Interactions at Protein-Nucleic Acid Interfaces
- PMID: 32845610
- PMCID: PMC7497705
- DOI: 10.1021/acs.accounts.0c00212
Dynamics of Ionic Interactions at Protein-Nucleic Acid Interfaces
Abstract
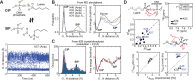
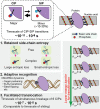
Molecular association of proteins with nucleic acids is required for many biological processes essential to life. Electrostatic interactions via ion pairs (salt bridges) of nucleic acid phosphates and protein side chains are crucial for proteins to bind to DNA or RNA. Counterions around the macromolecules are also key constituents for the thermodynamics of protein-nucleic acid association. Until recently, there had been only a limited amount of experiment-based information about how ions and ionic moieties behave in biological macromolecular processes. In the past decade, there has been significant progress in quantitative experimental research on ionic interactions with nucleic acids and their complexes with proteins. The highly negatively charged surfaces of DNA and RNA electrostatically attract and condense cations, creating a zone called the ion atmosphere. Recent experimental studies were able to examine and validate theoretical models on ions and their mobility and interactions with macromolecules. The ionic interactions are highly dynamic. The counterions rapidly diffuse within the ion atmosphere. Some of the ions are released from the ion atmosphere when proteins bind to nucleic acids, balancing the charge via intermolecular ion pairs of positively charged side chains and negatively charged backbone phosphates. Previously, the release of counterions had been implicated indirectly by the salt-concentration dependence of the association constant.Recently, direct detection of counterion release by NMR spectroscopy has become possible and enabled more accurate and quantitative analysis of the counterion release and its entropic impact on the thermodynamics of protein-nucleic acid association. Recent studies also revealed the dynamic nature of ion pairs of protein side chains and nucleic acid phosphates. These ion pairs undergo transitions between two major states. In one of the major states, the cation and the anion are in direct contact and form hydrogen bonds. In the other major state, the cation and the anion are separated by water. Transitions between these states rapidly occur on a picosecond to nanosecond time scale. When proteins interact with nucleic acids, interfacial arginine (Arg) and lysine (Lys) side chains exhibit considerably different behaviors. Arg side chains show a higher propensity to form rigid contacts with nucleotide bases, whereas Lys side chains tend to be more mobile at the molecular interfaces. The dynamic ionic interactions may facilitate adaptive molecular recognition and play both thermodynamic and kinetic roles in protein-nucleic acid interactions.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
Physicochemical Properties of Ion Pairs of Biological Macromolecules.Biomolecules. 2015 Sep 30;5(4):2435-63. doi: 10.3390/biom5042435. Biomolecules. 2015. PMID: 26437440 Free PMC article. Review.
-
NMR Methods for Characterizing the Basic Side Chains of Proteins: Electrostatic Interactions, Hydrogen Bonds, and Conformational Dynamics.Methods Enzymol. 2019;615:285-332. doi: 10.1016/bs.mie.2018.08.017. Epub 2018 Sep 27. Methods Enzymol. 2019. PMID: 30638532 Free PMC article. Review.
-
Changes in conformational dynamics of basic side chains upon protein-DNA association.Nucleic Acids Res. 2016 Aug 19;44(14):6961-70. doi: 10.1093/nar/gkw531. Epub 2016 Jun 10. Nucleic Acids Res. 2016. PMID: 27288446 Free PMC article.
-
Dynamic Equilibria of Short-Range Electrostatic Interactions at Molecular Interfaces of Protein-DNA Complexes.J Phys Chem Lett. 2015 Jul 16;6(14):2733-7. doi: 10.1021/acs.jpclett.5b01134. J Phys Chem Lett. 2015. PMID: 26207171 Free PMC article.
-
Statistical analysis of atomic contacts at RNA-protein interfaces.J Mol Recognit. 2001 Jul-Aug;14(4):199-214. doi: 10.1002/jmr.534. J Mol Recognit. 2001. PMID: 11500966
Cited by
-
Direct measurements of biomolecular electrostatics through experiments.Curr Opin Struct Biol. 2023 Oct;82:102680. doi: 10.1016/j.sbi.2023.102680. Epub 2023 Aug 11. Curr Opin Struct Biol. 2023. PMID: 37573815 Free PMC article. Review.
-
Exploring the bistable equilibrium of methylated CpG DNA recognition by the MBD2 protein.bioRxiv [Preprint]. 2025 Jun 30:2025.06.30.662303. doi: 10.1101/2025.06.30.662303. bioRxiv. 2025. PMID: 40631181 Free PMC article. Preprint.
-
Dynamics of Cations around DNA and Protein as Revealed by 23Na Diffusion NMR Spectroscopy.Anal Chem. 2022 Feb 8;94(5):2444-2452. doi: 10.1021/acs.analchem.1c04197. Epub 2022 Jan 26. Anal Chem. 2022. PMID: 35080384 Free PMC article.
-
Dynamic Autoinhibition of the HMGB1 Protein via Electrostatic Fuzzy Interactions of Intrinsically Disordered Regions.J Mol Biol. 2021 Sep 3;433(18):167122. doi: 10.1016/j.jmb.2021.167122. Epub 2021 Jun 25. J Mol Biol. 2021. PMID: 34181980 Free PMC article.
-
De novo determination of near-surface electrostatic potentials by NMR.Proc Natl Acad Sci U S A. 2021 Jun 22;118(25):e2104020118. doi: 10.1073/pnas.2104020118. Proc Natl Acad Sci U S A. 2021. PMID: 34161285 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

