Germline IKAROS dimerization haploinsufficiency causes hematologic cytopenias and malignancies
- PMID: 32845957
- PMCID: PMC7819759
- DOI: 10.1182/blood.2020007292
Germline IKAROS dimerization haploinsufficiency causes hematologic cytopenias and malignancies
Abstract
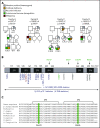
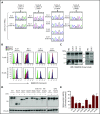
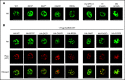
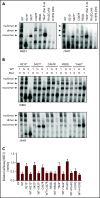
IKAROS is a transcription factor forming homo- and heterodimers and regulating lymphocyte development and function. Germline mutations affecting the IKAROS N-terminal DNA binding domain, acting in a haploinsufficient or dominant-negative manner, cause immunodeficiency. Herein, we describe 4 germline heterozygous IKAROS variants affecting its C-terminal dimerization domain, via haploinsufficiency, in 4 unrelated families. Index patients presented with hematologic disease consisting of cytopenias (thrombocytopenia, anemia, neutropenia)/Evans syndrome and malignancies (T-cell acute lymphoblastic leukemia, Burkitt lymphoma). These dimerization defective mutants disrupt homo- and heterodimerization in a complete or partial manner, but they do not affect the wild-type allele function. Moreover, they alter key mechanisms of IKAROS gene regulation, including sumoylation, protein stability, and the recruitment of the nucleosome remodeling and deacetylase complex; none affected in N-terminal DNA binding defects. These C-terminal dimerization mutations are largely associated with hematologic disorders, display dimerization haploinsufficiency and incomplete clinical penetrance, and differ from previously reported allelic variants in their mechanism of action. Dimerization mutants contribute to the growing spectrum of IKAROS-associated diseases displaying a genotype-phenotype correlation.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
-
- Georgopoulos K, Bigby M, Wang JH, et al. . The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79(1):143-156. - PubMed
-
- Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and Ikaros. Nat Rev Immunol. 2002;2(3):162-174. - PubMed
-
- Koipally J, Georgopoulos K. A molecular dissection of the repression circuitry of Ikaros. J Biol Chem. 2002;277(31):27697-27705. - PubMed
-
- Kim J, Sif S, Jones B, et al. . Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345-355. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

