Roadblocks and fast tracks: How RNA binding proteins affect the viral RNA journey in the cell
- PMID: 32847707
- PMCID: PMC7443355
- DOI: 10.1016/j.semcdb.2020.08.006
Roadblocks and fast tracks: How RNA binding proteins affect the viral RNA journey in the cell
Abstract
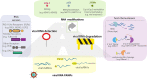
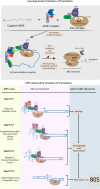
As obligate intracellular parasites with limited coding capacity, RNA viruses rely on host cells to complete their multiplication cycle. Viral RNAs (vRNAs) are central to infection. They carry all the necessary information for a virus to synthesize its proteins, replicate and spread and could also play essential non-coding roles. Regardless of its origin or tropism, vRNA has by definition evolved in the presence of host RNA Binding Proteins (RBPs), which resulted in intricate and complicated interactions with these factors. While on one hand some host RBPs recognize vRNA as non-self and mobilize host antiviral defenses, vRNA must also co-opt other host RBPs to promote viral infection. Focusing on pathogenic RNA viruses, we will review important scenarios of RBP-vRNA interactions during which host RBPs recognize, modify or degrade vRNAs. We will then focus on how vRNA hijacks the largest ribonucleoprotein complex (RNP) in the cell, the ribosome, to selectively promote the synthesis of its proteins. We will finally reflect on how novel technologies are helping in deepening our understanding of vRNA-host RBPs interactions, which can be ultimately leveraged to combat everlasting viral threats.
Keywords: Innate Immunity; RNA biology; RNA viruses; Technology; Viral RNA sensing; Viral Translation; Virology.
Copyright © 2020. Published by Elsevier Ltd.
Conflict of interest statement
The authors report no declarations of interest.
Figures

References
-
- Marraffini L.A. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

