Heterotypic cell-cell communication regulates glandular stem cell multipotency
- PMID: 32848220
- PMCID: PMC7116172
- DOI: 10.1038/s41586-020-2632-y
Heterotypic cell-cell communication regulates glandular stem cell multipotency
Abstract
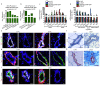
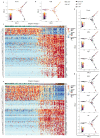
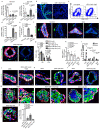
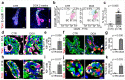
Glandular epithelia, including the mammary and prostate glands, are composed of basal cells (BCs) and luminal cells (LCs)1,2. Many glandular epithelia develop from multipotent basal stem cells (BSCs) that are replaced in adult life by distinct pools of unipotent stem cells1,3-8. However, adult unipotent BSCs can reactivate multipotency under regenerative conditions and upon oncogene expression3,9-13. This suggests that an active mechanism restricts BSC multipotency under normal physiological conditions, although the nature of this mechanism is unknown. Here we show that the ablation of LCs reactivates the multipotency of BSCs from multiple epithelia both in vivo in mice and in vitro in organoids. Bulk and single-cell RNA sequencing revealed that, after LC ablation, BSCs activate a hybrid basal and luminal cell differentiation program before giving rise to LCs-reminiscent of the genetic program that regulates multipotency during embryonic development7. By predicting ligand-receptor pairs from single-cell data14, we find that TNF-which is secreted by LCs-restricts BC multipotency under normal physiological conditions. By contrast, the Notch, Wnt and EGFR pathways were activated in BSCs and their progeny after LC ablation; blocking these pathways, or stimulating the TNF pathway, inhibited regeneration-induced BC multipotency. Our study demonstrates that heterotypic communication between LCs and BCs is essential to maintain lineage fidelity in glandular epithelial stem cells.
Conflict of interest statement
Figures
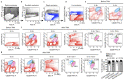
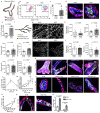
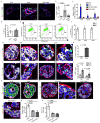
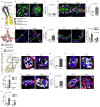



References
-
- Lloyd-Lewis B, Harris OB, Watson CJ, Davis FM. Mammary stem cells: premise, properties, and perspectives. Trends Cell Biol. 2017;27:556–567. - PubMed
-
- Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. - PubMed
-
- Ousset M, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat Cell Biol. 2012;14:1131–1138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

