Distinct viral reservoirs in individuals with spontaneous control of HIV-1
- PMID: 32848246
- PMCID: PMC7837306
- DOI: 10.1038/s41586-020-2651-8
Distinct viral reservoirs in individuals with spontaneous control of HIV-1
Abstract
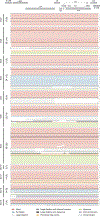
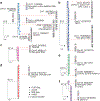
Sustained, drug-free control of HIV-1 replication is naturally achieved in less than 0.5% of infected individuals (here termed 'elite controllers'), despite the presence of a replication-competent viral reservoir1. Inducing such an ability to spontaneously maintain undetectable plasma viraemia is a major objective of HIV-1 cure research, but the characteristics of proviral reservoirs in elite controllers remain to be determined. Here, using next-generation sequencing of near-full-length single HIV-1 genomes and corresponding chromosomal integration sites, we show that the proviral reservoirs of elite controllers frequently consist of oligoclonal to near-monoclonal clusters of intact proviral sequences. In contrast to individuals treated with long-term antiretroviral therapy, intact proviral sequences from elite controllers were integrated at highly distinct sites in the human genome and were preferentially located in centromeric satellite DNA or in Krüppel-associated box domain-containing zinc finger genes on chromosome 19, both of which are associated with heterochromatin features. Moreover, the integration sites of intact proviral sequences from elite controllers showed an increased distance to transcriptional start sites and accessible chromatin of the host genome and were enriched in repressive chromatin marks. These data suggest that a distinct configuration of the proviral reservoir represents a structural correlate of natural viral control, and that the quality, rather than the quantity, of viral reservoirs can be an important distinguishing feature for a functional cure of HIV-1 infection. Moreover, in one elite controller, we were unable to detect intact proviral sequences despite analysing more than 1.5 billion peripheral blood mononuclear cells, which raises the possibility that a sterilizing cure of HIV-1 infection, which has previously been observed only following allogeneic haematopoietic stem cell transplantation2,3, may be feasible in rare instances.
Conflict of interest statement
ANE has received fees from ViiV Healthcare Co. within the past year for work unrelated to this project. All other authors declare that conflicts of interest do not exist.
Figures




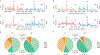


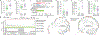

Comment in
-
HIV enters deep sleep in people who naturally control the virus.Nature. 2020 Sep;585(7824):190-191. doi: 10.1038/d41586-020-02438-7. Nature. 2020. PMID: 32848236 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- UM1 AI126620/AI/NIAID NIH HHS/United States
- U24 AI143502/AI/NIAID NIH HHS/United States
- R01 AI078799/AI/NIAID NIH HHS/United States
- R01 HL134539/HL/NHLBI NIH HHS/United States
- T32 AI060530/AI/NIAID NIH HHS/United States
- R33 DA047034/DA/NIDA NIH HHS/United States
- R01 AI130005/AI/NIAID NIH HHS/United States
- R33 AI116228/AI/NIAID NIH HHS/United States
- AI117841/NH/NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- AI052014/NH/NIH HHS/United States
- AI120008/NH/NIH HHS/United States
- P30 AI060354/AI/NIAID NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- AI135940/NH/NIH HHS/United States
- T32 AI007386/AI/NIAID NIH HHS/United States
- U01 AI135940/AI/NIAID NIH HHS/United States
- AI098487/NH/NIH HHS/United States
- P30 AI094189/AI/NIAID NIH HHS/United States
- U19 AI096109/AI/NIAID NIH HHS/United States
- U01 AI114235/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI098487/AI/NIAID NIH HHS/United States
- UM1 AI126603/AI/NIAID NIH HHS/United States
- R44 AI124996/AI/NIAID NIH HHS/United States
- DK120387/NH/NIH HHS/United States
- AI114235/NH/NIH HHS/United States
- R37 AI067073/AI/NIAID NIH HHS/United States
- R61 DA047034/DA/NIDA NIH HHS/United States
- U01 AI117841/AI/NIAID NIH HHS/United States
- R01 DK120387/DK/NIDDK NIH HHS/United States
- AI078799/NH/NIH HHS/United States
- DA047034/NH/NIH HHS/United States
- UM1 AI126611/AI/NIAID NIH HHS/United States
- R01 AI052014/AI/NIAID NIH HHS/United States
- R01 AI120008/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HL134539/NH/NIH HHS/United States
- AI116228/NH/NIH HHS/United States
- R21 AI116228/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

