LINC00662 Long Non-Coding RNA Knockdown Attenuates the Proliferation, Migration, and Invasion of Osteosarcoma Cells by Regulating the microRNA-15a-5p/Notch2 Axis
- PMID: 32848412
- PMCID: PMC7429411
- DOI: 10.2147/OTT.S256464
LINC00662 Long Non-Coding RNA Knockdown Attenuates the Proliferation, Migration, and Invasion of Osteosarcoma Cells by Regulating the microRNA-15a-5p/Notch2 Axis
Abstract
Purpose: Osteosarcoma (OS) is a frequently occurring malignancy in children and adolescents. In this study, we aimed to investigate the effects of the long non-coding RNA (lncRNA) LINC00662 (LINC00662) in OS and the underlying molecular mechanism.
Methods: The expression of LINC00662, microRNA-15a-5p (miR-15a-5p), and Notch2 in OS was detected by quantitative real-time polymerase chain reaction (qRT-PCR). The proliferation, migration, and invasion of OS cells were analyzed by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), wound-healing, and transwell assay. The interactions among LINC00662, miR-15a-5p, and Notch2 were determined by dual-luciferase reporter assays. A tumor xenograft model was established in mice for evaluating tumor growth in vivo.
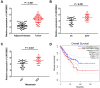
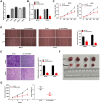
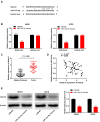
Results: The expression of LINC00662 and Notch2 was found to be upregulated in OS, but the expression of miR-15a-5p was downregulated. The results demonstrated that LINC00662 knockdown attenuated the proliferation, migration, and invasion of OS cells and suppressed tumor growth in mice. The study further demonstrated that LINC00662 directly interacted with miR-15a-5p, and that Notch2 was a target of miR-15a-5p. The inhibition of miR-15a-5p or Notch2 overexpression markedly reversed the suppressive effect of sh-LINC00662 on the proliferation, migration, and invasion of OS cells.
Conclusion: The study demonstrated that LINC00662 could be a potential biomarker for OS therapy, and LINC00662 knockdown suppressed the proliferation, migration, and invasion of OS cells by regulating the miR-15a-5p/Notch2 axis.
Keywords: Notch2; long non-coding RNA LINC00662; microRNA-15a-5p; osteosarcoma; proliferation.
© 2020 Liu and Meng.
Conflict of interest statement
The Authors declare that they have no conflicts of interest to disclose.
Figures



Similar articles
-
Knockdown of lncRNA LINC00662 suppresses malignant behaviour of osteosarcoma cells via competition with miR-30b-3p to regulate ELK1 expression.J Orthop Surg Res. 2022 Feb 5;17(1):74. doi: 10.1186/s13018-022-02964-2. J Orthop Surg Res. 2022. PMID: 35123530 Free PMC article.
-
LINC00662 triggers malignant progression of chordoma by the activation of RNF144B via targeting miR-16-5p.Eur Rev Med Pharmacol Sci. 2020 Feb;24(3):1007-1022. doi: 10.26355/eurrev_202002_20151. Eur Rev Med Pharmacol Sci. 2020. PMID: 32096180
-
Long non-coding RNA LINC00662 promotes proliferation and migration of breast cancer cells via regulating the miR-497-5p/EglN2 axis.Acta Biochim Pol. 2020 Jun 19;67(2):229-237. doi: 10.18388/abp.2020_5203. Acta Biochim Pol. 2020. PMID: 32558530
-
Long non-coding RNA small nucleolar RNA host gene 1 knockdown suppresses the proliferation, migration and invasion of osteosarcoma cells by regulating microRNA-424-5p/FGF2 in vitro.Exp Ther Med. 2021 Apr;21(4):325. doi: 10.3892/etm.2021.9756. Epub 2021 Feb 5. Exp Ther Med. 2021. PMID: 33732298 Free PMC article.
-
LncRNA LINC00662 Exerts an Oncogenic Effect on Osteosarcoma by the miR-16-5p/ITPR1 Axis.J Oncol. 2021 Sep 28;2021:8493431. doi: 10.1155/2021/8493431. eCollection 2021. J Oncol. 2021. PMID: 34621314 Free PMC article.
Cited by
-
A pan-cancer analysis of the prognostic value of long non-coding RNA LINC00662 in human cancers.Front Genet. 2022 Dec 8;13:1063119. doi: 10.3389/fgene.2022.1063119. eCollection 2022. Front Genet. 2022. PMID: 36568401 Free PMC article.
-
Knockdown of lncRNA LINC00662 suppresses malignant behaviour of osteosarcoma cells via competition with miR-30b-3p to regulate ELK1 expression.J Orthop Surg Res. 2022 Feb 5;17(1):74. doi: 10.1186/s13018-022-02964-2. J Orthop Surg Res. 2022. PMID: 35123530 Free PMC article.
-
Single-Cell Profiling of Tumor Microenvironment Heterogeneity in Osteosarcoma Identifies a Highly Invasive Subcluster for Predicting Prognosis.Front Oncol. 2022 Apr 6;12:732862. doi: 10.3389/fonc.2022.732862. eCollection 2022. Front Oncol. 2022. PMID: 35463309 Free PMC article.
-
Long Noncoding RNA MIR100HG Knockdown Attenuates Hepatocellular Carcinoma Progression by Regulating MicroRNA-146b-5p/Chromobox 6.Gastroenterol Res Pract. 2021 Jul 30;2021:6832518. doi: 10.1155/2021/6832518. eCollection 2021. Gastroenterol Res Pract. 2021. PMID: 34381502 Free PMC article.
-
Osteosarcoma in a ceRNET perspective.J Biomed Sci. 2024 Jun 5;31(1):59. doi: 10.1186/s12929-024-01049-y. J Biomed Sci. 2024. PMID: 38835012 Free PMC article. Review.
References
-
- Franceschini N, Cleton-Jansen A-M, Bovée JV. Bone: conventional osteosarcoma. Atlas Genet Cytogenet Oncol Haematol. 2019;(8). doi:10.4267/2042/70484. - DOI
-
- Wahidah T, Khattak MN, Wan-Arfah N, Naing NN. Five-year survival of osteosarcoma patients in Hospital Universiti Sains Malaysia (HUSM): an eleven year review. Res J Pharm Technol. 2018;11(8):3534–3542. doi:10.5958/0974-360X.2018.00653.4 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

