Identification of Potential Interacting Proteins With the Extracellular Loops of the Neuronal Glycoprotein M6a by TMT/MS
- PMID: 32848694
- PMCID: PMC7396582
- DOI: 10.3389/fnsyn.2020.00028
Identification of Potential Interacting Proteins With the Extracellular Loops of the Neuronal Glycoprotein M6a by TMT/MS
Abstract
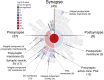
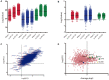
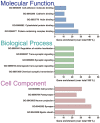
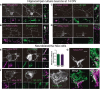
Nowadays, great efforts are made to gain insight into the molecular mechanisms that underlie structural neuronal plasticity. Moreover, the identification of signaling pathways involved in the development of psychiatric disorders aids the screening of possible therapeutic targets. Genetic variations or alterations in GPM6A expression are linked to neurological disorders such as schizophrenia, depression, and Alzheimer's disease. GPM6A encodes the neuronal surface glycoprotein M6a that promotes filopodia/spine, dendrite, and synapse formation by unknown mechanisms. A substantial body of evidence suggests that the extracellular loops of M6a command its function. However, the proteins that associate with them and that modulate neuronal plasticity have not been determined yet. To address this question, we generated a chimera protein that only contains the extracellular loops of M6a and performed a co-immunoprecipitation with rat hippocampus samples followed by TMT/MS. Here, we report 72 proteins, which are good candidates to interact with M6a's extracellular loops and modify its function. Gene ontology (GO) analysis showed that 63% of the potential M6a's interactor proteins belong to the category "synapse," at both sides of the synaptic cleft, "neuron projections" (51%) and "presynapse" (49%). In this sense, we showed that endogenous M6a interacts with piccolo, synaptic vesicle protein 2B, and synapsin 1 in mature cultured hippocampal neurons. Interestingly, about 28% of the proteins left were related to the "myelin sheath" annotation, suggesting that M6a could interact with proteins at the surface of oligodendrocytes. Indeed, we demonstrated the (cis and trans) interaction between M6a and proteolipid protein (PLP) in neuroblastoma N2a cells. Finally, the 72 proteins were subjected to disease-associated genes and variants screening by DisGeNET. Apart from the diseases that have already been associated with M6a, most of the proteins are also involved in "autistic disorder," "epilepsy," and "seizures" increasing the spectrum of disorders in which M6a could play a role. Data are available via ProteomeXchange with identifier PXD017347.
Keywords: mass spectometry; protein-protein interaction; proteolipd protein family; rat hippocampal neurons; synaptic proteins.
Copyright © 2020 Aparicio, Formoso, León, Frasch and Scorticati.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

