The σ Subunit-Remodeling Factors: An Emerging Paradigms of Transcription Regulation
- PMID: 32849409
- PMCID: PMC7403470
- DOI: 10.3389/fmicb.2020.01798
The σ Subunit-Remodeling Factors: An Emerging Paradigms of Transcription Regulation
Abstract
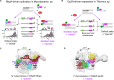
Transcription initiation is a key checkpoint and highly regulated step of gene expression. The sigma (σ) subunit of RNA polymerase (RNAP) controls all transcription initiation steps, from recognition of the -10/-35 promoter elements, upon formation of the closed promoter complex (RPc), to stabilization of the open promoter complex (RPo) and stimulation of the primary steps in RNA synthesis. The canonical mechanism to regulate σ activity upon transcription initiation relies on activators that recognize specific DNA motifs and recruit RNAP to promoters. This mini-review describes an emerging group of transcriptional regulators that form a complex with σ or/and RNAP prior to promoter binding, remodel the σ subunit conformation, and thus modify RNAP activity. Such strategy is widely used by bacteriophages to appropriate the host RNAP. Recent findings on RNAP-binding protein A (RbpA) from Mycobacterium tuberculosis and Crl from Escherichia coli suggest that activator-driven changes in σ conformation can be a widespread regulatory mechanism in bacteria.
Keywords: RNAP-binding transcriptional regulators; RbpA; Tuberculosis; promoter specificity; sigma subunit conformational dynamics.
Copyright © 2020 Vishwakarma and Brodolin.
Figures

References
-
- Banta A. B., Chumanov R. S., Yuan A. H., Lin H., Campbell E. A., Burgess R. R., et al. (2013). Key features of σS required for specific recognition by Crl, a transcription factor promoting assembly of RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 110 15955–16960. 10.1073/pnas.1311642110 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

