Characterization of the Bronchoalveolar Lavage Fluid by Single Cell Gene Expression Analysis in Healthy Dogs: A Promising Technique
- PMID: 32849601
- PMCID: PMC7406785
- DOI: 10.3389/fimmu.2020.01707
Characterization of the Bronchoalveolar Lavage Fluid by Single Cell Gene Expression Analysis in Healthy Dogs: A Promising Technique
Abstract
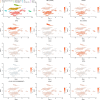
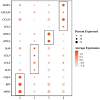
Single-cell mRNA-sequencing (scRNA-seq) is a technique which enables unbiased, high throughput and high-resolution transcriptomic analysis of the heterogeneity of cells within a population. This recent technique has been described in humans, mice and other species in various conditions to cluster cells in populations and identify new subpopulations, as well as to study the gene expression of cells in various tissues, conditions and origins. In dogs, a species for which markers of cell populations are often limiting, scRNA-seq presents with elevated yet untested potential for the study of tissue composition. As a proof of principle, we used scRNA-seq to identify cellular populations of the bronchoalveolar lavage fluid (BALF) in healthy dogs (n = 4). A total of 5,710 cells were obtained and analyzed by scRNA-seq. Fourteen distinct clusters of cells were identified, further identified as macrophages/monocytes (4 clusters), T cells (2 clusters) and B cells (1 cluster), neutrophils (1 cluster), mast cells (1 cluster), mature or immature dendritic cells (1 cluster each), ciliated or non-ciliated epithelial cells (1 cluster each) and cycling cells (1 cluster). We used for the first time in dogs the scRNA-seq to investigate cellular subpopulations of the BALF of dog. This study hence expands our knowledge on dog lung immune cell populations, paves the way for the investigation at single-cell level of lower respiratory diseases in dogs, and establishes that scRNA-seq is a powerful tool for the study of dog tissue composition.
Keywords: bronchoalveolar lavage fluid; cell; dog; lung; single-cell RNA-sequencing.
Copyright © 2020 Fastrès, Pirottin, Fievez, Marichal, Desmet, Bureau and Clercx.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

