Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment
- PMID: 32849616
- PMCID: PMC7417513
- DOI: 10.3389/fimmu.2020.01731
Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment
Abstract
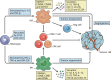
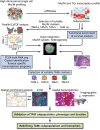
The immunosuppressive status of the tumor microenvironment (TME) remains poorly defined due to a lack of understanding regarding the function of tumor-associated macrophages (TAMs), which are abundant in the TME. TAMs are crucial drivers of tumor progression, metastasis, and resistance to therapy. Intra- and inter-tumoral spatial heterogeneities are potential keys to understanding the relationships between subpopulations of TAMs and their functions. Antitumor M1-like and pro-tumor M2-like TAMs coexist within tumors, and the opposing effects of these M1/M2 subpopulations on tumors directly impact current strategies to improve antitumor immune responses. Recent studies have found significant differences among monocytes or macrophages from distinct tumors, and other investigations have explored the existence of diverse TAM subsets at the molecular level. In this review, we discuss emerging evidence highlighting the redefinition of TAM subpopulations and functions in the TME and the possibility of separating macrophage subsets with distinct functions into antitumor M1-like and pro-tumor M2-like TAMs during the development of tumors. Such redefinition may relate to the differential cellular origin and monocyte and macrophage plasticity or heterogeneity of TAMs, which all potentially impact macrophage biomarkers and our understanding of how the phenotypes of TAMs are dictated by their ontogeny, activation status, and localization. Therefore, the detailed landscape of TAMs must be deciphered with the integration of new technologies, such as multiplexed immunohistochemistry (mIHC), mass cytometry by time-of-flight (CyTOF), single-cell RNA-seq (scRNA-seq), spatial transcriptomics, and systems biology approaches, for analyses of the TME.
Keywords: multiplexed immunohistochemical staining; single-cell sequencing; spatial transcriptomics; tumor microenvironment; tumor-associated macrophages.
Copyright © 2020 Wu, Lin, Li, Yuan, Xu, Ni and Xu.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

