Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past
- PMID: 32849654
- PMCID: PMC7426442
- DOI: 10.3389/fimmu.2020.01949
Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past
Abstract
After the 1918 flu pandemic, the world is again facing a similar situation. However, the advancement in medical science has made it possible to identify that the novel infectious agent is from the coronavirus family. Rapid genome sequencing by various groups helped in identifying the structure and function of the virus, its immunogenicity in diverse populations, and potential preventive measures. Coronavirus attacks the respiratory system, causing pneumonia and lymphopenia in infected individuals. Viral components like spike and nucleocapsid proteins trigger an immune response in the host to eliminate the virus. These viral antigens can be either recognized by the B cells or presented by MHC complexes to the T cells, resulting in antibody production, increased cytokine secretion, and cytolytic activity in the acute phase of infection. Genetic polymorphism in MHC enables it to present some of the T cell epitopes very well over the other MHC alleles. The association of MHC alleles and its downregulated expression has been correlated with disease severity against influenza and coronaviruses. Studies have reported that infected individuals can, after recovery, induce strong protective responses by generating a memory T-cell pool against SARS-CoV and MERS-CoV. These memory T cells were not persistent in the long term and, upon reactivation, caused local damage due to cross-reactivity. So far, the reports suggest that SARS-CoV-2, which is highly contagious, shows related symptoms in three different stages and develops an exhaustive T-cell pool at higher loads of viral infection. As there are no specific treatments available for this novel coronavirus, numerous small molecular drugs that are being used for the treatment of diseases like SARS, MERS, HIV, ebola, malaria, and tuberculosis are being given to COVID-19 patients, and clinical trials for many such drugs have already begun. A classical immunotherapy of convalescent plasma transfusion from recovered patients has also been initiated for the neutralization of viremia in terminally ill COVID-19 patients. Due to the limitations of plasma transfusion, researchers are now focusing on developing neutralizing antibodies against virus particles along with immuno-modulation of cytokines like IL-6, Type I interferons (IFNs), and TNF-α that could help in combating the infection. This review highlights the similarities of the coronaviruses that caused SARS and MERS to the novel SARS-CoV-2 in relation to their pathogenicity and immunogenicity and also focuses on various treatment strategies that could be employed for curing COVID-19.
Keywords: COVID-19; HLA; MHC presentation; T cells; coronavirus; immune response; memory T cell.
Copyright © 2020 Shah, Firmal, Alam, Ganguly and Chattopadhyay.
Figures
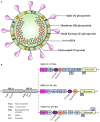


References
-
- Ong SWX, Tan YK, Chia PY, Lee TH, Ng OT, Wong MSY, et al. . Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. JAMA - J Am Med Assoc. (2020) 323:5–7. 10.1001/jama.2020.3227 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

