Aphid-Responsive Defense Networks in Hybrid Switchgrass
- PMID: 32849703
- PMCID: PMC7412557
- DOI: 10.3389/fpls.2020.01145
Aphid-Responsive Defense Networks in Hybrid Switchgrass
Abstract
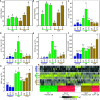
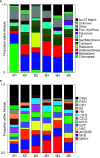
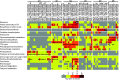
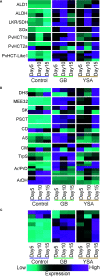
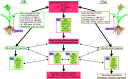
Aphid herbivory elicits plant defense-related networks that are influenced by host genetics. Plants of the upland switchgrass (Panicum virgatum) cultivar Summer can be a suitable host for greenbug aphids (Schizaphis graminum; GB), and yellow sugarcane aphids (Sipha flava, YSA), whereas the lowland cultivar Kanlow exhibited multi-species resistance that curtails aphid reproduction. However, stabilized hybrids of Summer (♀) x Kanlow (♂) (SxK) with improved agronomics can be damaged by both aphids. Here, hormone and metabolite analyses, coupled with RNA-Seq analysis of plant transcriptomes, were utilized to delineate defense networks induced by aphid feeding in SxK switchgrass and pinpoint plant transcription factors (TFs), such as WRKYs that potentially regulate these responses. Abscisic acid (ABA) levels were significantly higher in GB infested plants at 5 and 10 days after infestation (DAI). ABA levels were highest at 15DAI in YSA infested plants. Jasmonic acid levels were significantly elevated under GB infestation, while salicylic acid levels were signifi40cantly elevated only at 15 DAI in YSA infested plants. Similarly, levels of several metabolites were altered in common or specifically to each aphid. YSA infestation induced a significant enrichment of flavonoids consistent with an upregulation of many genes associated with flavonoid biosynthesis at 15DAI. Gene co-expression modules that responded singly to either aphid or in common to both aphids were differentiated and linked to specific TFs. Together, these data provide important clues into the interplay of metabolism and transcriptional remodeling accompanying defense responses to aphid herbivory in hybrid switchgrass.
Keywords: Panicum virgatum; aphids; gene-networks; hybrid switchgrass; metabolites; plant defense; transcription factors; transcriptomes.
Copyright © 2020 Koch, Palmer, Donze-Reiner, Scully, Seravalli, Amundsen, Twigg, Louis, Bradshaw, Heng-Moss and Sarath.
Figures


References
LinkOut - more resources
Full Text Sources

