Genome-Wide Transcriptional Analysis Reveals Alternative Splicing Event Profiles in Hepatocellular Carcinoma and Their Prognostic Significance
- PMID: 32849842
- PMCID: PMC7432180
- DOI: 10.3389/fgene.2020.00879
Genome-Wide Transcriptional Analysis Reveals Alternative Splicing Event Profiles in Hepatocellular Carcinoma and Their Prognostic Significance
Abstract
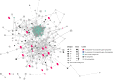
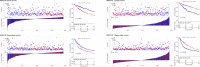
Accumulating evidence indicates an unexpected role of aberrant splicing in hepatocellular carcinoma (HCC) that has been seriously neglected in previous studies. There is a need for a detailed analysis of alternative splicing (AS) and its underlying biological and clinical relevance in HCC. In this study, clinical information and corresponding RNA sequencing data of HCC patients were obtained from The Cancer Genome Atlas. Percent spliced in (PSI) values and transcriptional splicing patterns of genes were determined from the original RNA sequencing data using SpliceSeq. Then, based on the PSI values of AS events in different patients, a series of bioinformatics methods was used to identify differentially expressed AS events (DEAS), determine potential regulatory relationships, and investigate the correlation between DEAS and the patients' clinicopathological features. Finally, 25,934 AS events originating from 8,795 genes were screened with high reliability; 263 of these AS events were identified as DEAS. The parent genes of these DEAS formed an intricate network with roles in the regulation of cancer-related pathway and liver metabolism. In HCC, 36 splicing factors were involved in the dysregulation of part DEAS, 100 DEAS events were correlated with overall survival, and 71 DEAS events were correlated with disease-free survival. Stratifying HCC patients according to DEAS resulted in four clusters with different survival patterns. Significant variations in AS occurred during HCC initiation and maintenance; these are likely to be vital both for biological processes and in prognosis. The HCC-related AS events identified here and the splicing networks constructed will be valuable in deciphering the underlying role of AS in HCC.
Keywords: RNA-seq; alternative splicing; genome-wide; hepatocellular carcinoma; prognosis.
Copyright © 2020 Xiong, Yang, Wang, Riaz, Xu, Lv, Zhou, Li, Li, Sun, Tao and Li.
Figures







References
LinkOut - more resources
Full Text Sources
Research Materials

