A New Method to Obtain the Complete Genome Sequence of Multiple-Component Circular ssDNA Viruses by Transcriptome Analysis
- PMID: 32850712
- PMCID: PMC7396673
- DOI: 10.3389/fbioe.2020.00832
A New Method to Obtain the Complete Genome Sequence of Multiple-Component Circular ssDNA Viruses by Transcriptome Analysis
Abstract
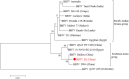
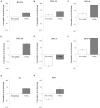
Circular single-stranded DNA (ssDNA) viruses are widely distributed globally, infecting diverse hosts ranging from bacteria, archaea, and eukaryotes. Among these, the genome of Banana bunchy top virus (BBTV) comprises at least six circular, ssDNA components that are ∼1 kb in length. Its genome is usually amplified and obtained at the DNA level. However, RNA-based techniques to obtain the genome sequence of such multi-component viruses have not been reported. In this study, transcriptome sequencing analysis showed that the full-length of BBTV each genomic component was transcribed into viral mRNA (vmRNA). Accordingly, the near-complete genome of BBTV B2 isolate was assembled using transcriptome sequencing data from virus-infected banana leaves. Assembly analysis of BBTV-derived reads indicated that the full-length sequences were obtained for DNA-R, DNA-U3, DNA-S, DNA-M, DNA-N, NewS2, and Sat4 components, while two gaps (73 and 25 nt) missing in the DNA-C component which was further filled by reverse transcription-PCR (RT-PCR). The RT-qPCR analysis indicated that the vmRNA levels of coding regions were 3.19-103.53 folds higher than those of non-coding regions, implying that the integrity of genome assembly depended on the transcription level of non-coding region. In conclusion, this study proposes a new approach to obtain the genome of nanovirids, and provides insights for studying the transcriptional mechanism of the family Nanoviridae, Genomoviridae, and Geminiviridae.
Keywords: DNA virus; genome assembly; multi-component; nanovirids; transcriptomic sequencing.
Copyright © 2020 Yu, Zhang, Wang and Liu.
Figures


Similar articles
-
Identification and characterization of a distinct banana bunchy top virus isolate of Pacific-Indian Oceans group from North-East India.Virus Res. 2014 Apr;183:41-9. doi: 10.1016/j.virusres.2014.01.017. Epub 2014 Jan 24. Virus Res. 2014. PMID: 24468493
-
Evidence of recombination in the Banana bunchy top virus genome.Infect Genet Evol. 2011 Aug;11(6):1293-300. doi: 10.1016/j.meegid.2011.04.015. Epub 2011 Apr 22. Infect Genet Evol. 2011. PMID: 21539936
-
Occurrence of Banana Bunchy Top Disease Caused by the Banana bunchy top virus on Banana and Plantain (Musa sp.) in Cameroon.Plant Dis. 2009 Oct;93(10):1076. doi: 10.1094/PDIS-93-10-1076C. Plant Dis. 2009. PMID: 30754355
-
Independent modulation of individual genomic component transcription and a cis-acting element related to high transcriptional activity in a multipartite DNA virus.BMC Genomics. 2019 Jul 11;20(1):573. doi: 10.1186/s12864-019-5901-0. BMC Genomics. 2019. PMID: 31296162 Free PMC article.
-
Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes.Ann N Y Acad Sci. 2015 Apr;1341:41-53. doi: 10.1111/nyas.12675. Epub 2015 Feb 11. Ann N Y Acad Sci. 2015. PMID: 25675979 Review.
References
-
- Amin I., Ilyas M., Qazi J., Bashir R., Yadav J. S., Mansoor S., et al. (2011). Identification of a major pathogenicity determinant and suppressors of RNA silencing encoded by a South Pacific isolate of Banana bunchy top virus originating from Pakistan. Virus Genes 42 272–281. 10.1007/s11262-010-0559-3 - DOI - PubMed
-
- Bashir R., Javed F., Ahmed R., Mansoor S. (2012). Use of rolling circle amplification for the identification of unknown components of Banana bunchy top virus from Pakistan. Pakistan J. Life Soc. Sci. 10 91–97.
-
- Briddon R. W., Martin D. P., Roumagnac P., Navas-Castillo J., Fiallo-Olivé E., Moriones E., et al. (2018). Alphasatellitidae: a new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch. Virol. 163 2587–2600. 10.1007/s00705-018-3854-2 - DOI - PubMed
-
- Dayaram A., Potter K. A., Pailes R., Marinov M., Rosenstein D. D., Varsani A. (2015). Identification of diverse circular single-stranded DNA viruses in adult dragonflies and damselflies (Insecta: Odonata) of Arizona and Oklahoma. USA. Infect. Genet. Evol. 30 278–287. 10.1016/j.meegid.2014.12.037 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

