Cell Reprogramming With CRISPR/Cas9 Based Transcriptional Regulation Systems
- PMID: 32850737
- PMCID: PMC7399070
- DOI: 10.3389/fbioe.2020.00882
Cell Reprogramming With CRISPR/Cas9 Based Transcriptional Regulation Systems
Abstract
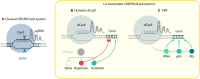
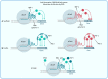
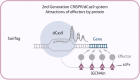
The speed of reprogramming technologies evolution is rising dramatically in modern science. Both the scientific community and health workers depend on such developments due to the lack of safe autogenic cells and tissues for regenerative medicine, genome editing tools and reliable screening techniques. To perform experiments efficiently and to propel the fundamental science it is important to keep up with novel modifications and techniques that are being discovered almost weekly. One of them is CRISPR/Cas9 based genome and transcriptome editing. The aim of this article is to summarize currently existing CRISPR/Cas9 applications for cell reprogramming, mainly, to compare them with other non-CRISPR approaches and to highlight future perspectives and opportunities.
Keywords: CRISPR/Cas9; dCas9; genome screening; human cell reprogramming; regenerative medicine; transactivator systems.
Copyright © 2020 Shakirova, Ovchinnikova and Dashinimaev.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

