A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction
- PMID: 32850967
- PMCID: PMC7417479
- DOI: 10.3389/fmolb.2020.00179
A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction
Abstract
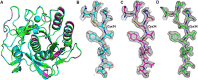
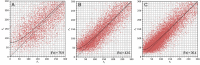
MicroED has recently emerged as a powerful method for the analysis of biological structures at atomic resolution. This technique has been largely limited to protein nanocrystals which grow either as needles or plates measuring only a few hundred nanometers in thickness. Furthermore, traditional microED data processing uses established X-ray crystallography software that is not optimized for handling compound effects that are unique to electron diffraction data. Here, we present an integrated workflow for microED, from sample preparation by cryo-focused ion beam milling, through data collection with a standard Ceta-D detector, to data processing using the DIALS software suite, thus enabling routine atomic structure determination of protein crystals of any size and shape using microED. We demonstrate the effectiveness of the workflow by determining the structure of proteinase K to 2.0 Å resolution and show the advantage of using protein crystal lamellae over nanocrystals.
Keywords: cryoEM; cryoFIB; crystallography; lamella; microED; nanocrystals; proteinase K.
Copyright © 2020 Beale, Waterman, Hecksel, van Rooyen, Gilchrist, Parkhurst, de Haas, Buijsse, Evans and Zhang.
Figures

References
-
- Arndt U. W., Wonacott A. J. (1977). The Rotation Method in Crystallography. Amsterdam: North-Holland Publishing Company.
-
- Clabbers M. T. B., Abrahams J. P. (2018). Electron diffraction and three-dimensional crystallography for structural biology. Crystallogr. Rev. 24 176–204. 10.1080/0889311X.2018.1446427 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

