Conjugative Coupling Proteins and the Role of Their Domains in Conjugation, Secondary Structure and in vivo Subcellular Location
- PMID: 32850972
- PMCID: PMC7431656
- DOI: 10.3389/fmolb.2020.00185
Conjugative Coupling Proteins and the Role of Their Domains in Conjugation, Secondary Structure and in vivo Subcellular Location
Abstract
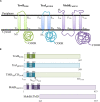
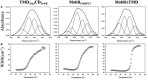
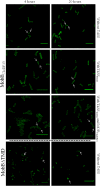
Type IV Coupling Proteins (T4CPs) are essential elements in many type IV secretion systems (T4SSs). The members of this family display sequence, length, and domain architecture heterogeneity, being the conserved Nucleotide-Binding Domain the motif that defines them. In addition, most T4CPs contain a Transmembrane Domain (TMD) in the amino end and an All-Alpha Domain facing the cytoplasm. Additionally, a few T4CPs present a variable domain at the carboxyl end. The structural paradigm of this family is TrwBR388, the T4CP of conjugative plasmid R388. This protein has been widely studied, in particular the role of the TMD on the different characteristics of TrwBR388. To gain knowledge about T4CPs and their TMD, in this work a chimeric protein containing the TMD of TraJpKM101 and the cytosolic domain of TrwBR388 has been constructed. Additionally, one of the few T4CPs of mobilizable plasmids, MobBCloDF13 of mobilizable plasmid CloDF13, together with its TMD-less mutant MobBΔTMD have been studied. Mating studies showed that the chimeric protein is functional in vivo and that it exerted negative dominance against the native proteins TrwBR388 and TraJpKM101. Also, it was observed that the TMD of MobBCloDF13 is essential for the mobilization of CloDF13 plasmid. Analysis of the secondary structure components showed that the presence of a heterologous TMD alters the structure of the cytosolic domain in the chimeric protein. On the contrary, the absence of the TMD in MobBCloDF13 does not affect the secondary structure of its cytosolic domain. Subcellular localization studies showed that T4CPs have a unipolar or bipolar location, which is enhanced by the presence of the remaining proteins of the conjugative system. Unlike what has been described for TrwBR388, the TMD is not an essential element for the polar location of MobBCloDF13. The main conclusion is that the characteristics described for the paradigmatic TrwBR388 T4CP should not be ascribed to the whole T4CP family. Specifically, it has been proven that the mobilizable plasmid-related MobBCloDF13 presents different characteristics regarding the role of its TMD. This work will contribute to better understand the T4CP family, a key element in bacterial conjugation, the main mechanism responsible for antibiotic resistance spread.
Keywords: antibiotic resistance spread; bacterial conjugation; coupling proteins; membrane proteins; type IV secretion systems.
Copyright © 2020 Álvarez-Rodríguez, Ugarte-Uribe, de la Arada, Arrondo, Garbisu and Alkorta.
Figures

References
-
- Andraka N., Sánchez-Magraner L., García-Pacios M., Goñi F. M., Arrondo J. L. R. (2017). The conformation of human phospholipid scramblase 1, as studied by infrared spectroscopy. Effects of calcium and detergent. Biochim. Biophys. Acta Biomembr. 1859 1019–1028. 10.1016/j.bbamem.2017.02.015 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

