Using Atomic Force Microscopy To Illuminate the Biophysical Properties of Microbes
- PMID: 32851362
- PMCID: PMC7447269
- DOI: 10.1021/acsabm.9b00973
Using Atomic Force Microscopy To Illuminate the Biophysical Properties of Microbes
Abstract
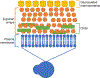
Since its invention in 1986, atomic force microscopy (AFM) has grown from a system designed for imaging inorganic surfaces to a tool used to probe the biophysical properties of living cells and tissues. AFM is a scanning probe technique and uses a pyramidal tip attached to a flexible cantilever to scan across a surface, producing a highly detailed image. While many research articles include AFM images, fewer include force-distance curves, from which several biophysical properties can be determined. In a single force-distance curve, the cantilever is lowered and raised from the surface, while the forces between the tip and the surface are monitored. Modern AFM has a wide variety of applications, but this review will focus on exploring the mechanobiology of microbes, which we believe is of particular interest to those studying biomaterials. We briefly discuss experimental design as well as different ways of extracting meaningful values related to cell surface elasticity, cell stiffness, and cell adhesion from force-distance curves. We also highlight both classic and recent experiments using AFM to illuminate microbial biophysical properties.
Keywords: adhesion; atomic force microscopy; cell stiffness; force–distance curve; membrane elasticity; microbes.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

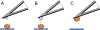
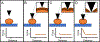

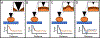


Similar articles
-
Atomic force microscopy (AFM).Curr Protoc Microbiol. 2008 Feb;Chapter 2:Unit 2C.2. doi: 10.1002/9780471729259.mc02c02s8. Curr Protoc Microbiol. 2008. PMID: 18770536
-
The application of atomic force microscopy in mineral flotation.Adv Colloid Interface Sci. 2018 Jun;256:373-392. doi: 10.1016/j.cis.2018.01.004. Epub 2018 Feb 6. Adv Colloid Interface Sci. 2018. PMID: 29559086 Review.
-
Evaluating the interaction of bacteria with biomaterials using atomic force microscopy.J Biomater Sci Polym Ed. 1998;9(12):1361-73. doi: 10.1163/156856298x00442. J Biomater Sci Polym Ed. 1998. PMID: 9860175
-
Multiparametric Atomic Force Microscopy Imaging of Biomolecular and Cellular Systems.Acc Chem Res. 2017 Apr 18;50(4):924-931. doi: 10.1021/acs.accounts.6b00638. Epub 2017 Mar 28. Acc Chem Res. 2017. PMID: 28350161 Review.
-
Analysis of bacterial adhesion using a gradient force analysis method and colloid probe atomic force microscopy.Langmuir. 2004 Sep 28;20(20):8817-22. doi: 10.1021/la0488203. Langmuir. 2004. PMID: 15379512
Cited by
-
Mechanobiology as a tool for addressing the genotype-to-phenotype problem in microbiology.Biophys Rev (Melville). 2023 May 12;4(2):021304. doi: 10.1063/5.0142121. eCollection 2023 Jun. Biophys Rev (Melville). 2023. PMID: 38504926 Free PMC article. Review.
-
Extracellular Polymeric Substance Protects Some Cells in an Escherichia coli Biofilm from the Biomechanical Consequences of Treatment with Magainin 2.Microorganisms. 2021 Apr 30;9(5):976. doi: 10.3390/microorganisms9050976. Microorganisms. 2021. PMID: 33946431 Free PMC article.
-
Pseudomonas aeruginosa assembles H1-T6SS in response to physical and chemical damage of the outer membrane.Sci Adv. 2025 Mar 7;11(10):eadr1713. doi: 10.1126/sciadv.adr1713. Epub 2025 Mar 5. Sci Adv. 2025. PMID: 40043119 Free PMC article.
-
Quantitative study of early-stage transient bacterial adhesion to bioactive glass and glass ceramics: atomic force microscopic observations.Sci Rep. 2024 Sep 2;14(1):20336. doi: 10.1038/s41598-024-67716-0. Sci Rep. 2024. PMID: 39223136 Free PMC article.
-
The intricate link between membrane lipid structure and composition and membrane structural properties in bacterial membranes.Chem Sci. 2024 Jan 31;15(10):3408-3427. doi: 10.1039/d3sc04523d. eCollection 2024 Mar 6. Chem Sci. 2024. PMID: 38455013 Free PMC article. Review.
References
-
- Binnig G; Quate C; Gerber C Atomic Force Microscope. Phys. Rev. Lett. 1986, 56 (9), 930–933. - PubMed
-
- Radmacher M; Tillamnn R; Fritz M; Gaub H From Molecules to Cells: Imaging Soft Samples with the Atomic Force Microscope. Science 1992, 257 (5078), 1900–1905. - PubMed
-
- Horber J;Miles M Scanning Probe Evolution in Biology. Science 2003, 302 (5647), 1002–1005. - PubMed
-
- Alessandrini A; Facci P AFM: A Versatile Tool in Biophysics. Meas. Sci. Technol. 2005, 16 (6), R65–R92.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
