In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly
- PMID: 32851912
- PMCID: PMC7484575
- DOI: 10.1080/07391102.2020.1804457
In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly
Abstract
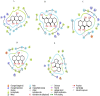
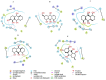
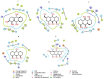
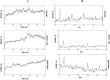
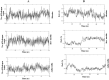
Currently, there is no specific treatment to cure COVID-19. Many medicinal plants have antiviral, antioxidant, antibacterial, antifungal, anticancer, wound healing etc. Therefore, the aim of the current study was to screen for potent inhibitors of N-terminal domain (NTD) of nucleocapsid phosphoprotein of SARS-CoV-2. The structure of NTD of RNA binding domain of nucleocapsid phosphoprotein of SARS coronavirus 2 was retrieved from the Protein Data Bank (PDB 6VYO) and the structures of 100 different phytocompounds were retrieved from Pubchem. The receptor protein and ligands were prepared using Schrodinger's Protein Preparation Wizard. Molecular docking was done by using the Schrodinger's maestro 12.0 software. Drug likeness and toxicity of active phytocompounds was predicted by using Swiss adme, admetSAR and protox II online servers. Molecular dynamic simulation of the best three protein- ligand complexes (alizarin, aloe-emodin and anthrarufin) was performed to study the interaction stability. We have identified three potential active sites (named as A, B, C) on receptor protein for efficient binding of the phytocompounds. We found that, among 100 phytocompounds, emodin, aloe-emodin, anthrarufin, alizarine, and dantron of Rheum emodi showed good binding affinity at all the three active sites of RNA binding domain of nucleocapsid phosphoprotein of COVID-19.The binding energies of emodin, aloe-emodin, anthrarufin, alizarine, and dantron were -8.299, -8.508, -8.456, -8.441, and -8.322 Kcal mol-1 respectively (site A), -7.714, -6.433, -6.354, -6.598, and -6.99 Kcal mol-1 respectively (site B), and -8.299, 8.508, 8.538, 8.841, and 8.322 Kcal mol-1 respectively (site C). All the active phytocompounds follows the drug likeness properties, non-carcinogenic, and non-toxic. Theses phytocompounds (alone or in combination) could be developed into effective therapy against COVID-19. From MD simulation data, we found that all three complexes of 6VYO with alizarin, aloe-emodin and anthrarufin were stable up to 50 ns. These phytocompounds can be tested further for in vitro or in vivo and used as a potential drug to cure SARS-CoV-2 infection.Communicated by Ramaswamy H. Sarma.
Keywords: Active site prediction and MD simulation; COVID-19; Molecular docking; Phytocompounds; RNA binding domain of nucleocapsid phosphoprotein; antiviral and toxicity.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures







Similar articles
-
Molecular docking studies of phytocompounds of Rheum emodi Wall with proteins responsible for antibiotic resistance in bacterial and fungal pathogens: in silico approach to enhance the bio-availability of antibiotics.J Biomol Struct Dyn. 2022 May;40(8):3789-3803. doi: 10.1080/07391102.2020.1850364. Epub 2020 Nov 23. J Biomol Struct Dyn. 2022. PMID: 33225862
-
Comprehensive Virtual Screening of the Antiviral Potentialities of Marine Polycyclic Guanidine Alkaloids against SARS-CoV-2 (COVID-19).Biomolecules. 2021 Mar 19;11(3):460. doi: 10.3390/biom11030460. Biomolecules. 2021. PMID: 33808721 Free PMC article.
-
Phytocompounds of Rheum emodi, Thymus serpyllum, and Artemisia annua Inhibit Spike Protein of SARS-CoV-2 Binding to ACE2 Receptor: In Silico Approach.Curr Pharmacol Rep. 2021;7(4):135-149. doi: 10.1007/s40495-021-00259-4. Epub 2021 Jul 15. Curr Pharmacol Rep. 2021. PMID: 34306988 Free PMC article. Review.
-
Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants - Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi) - a molecular docking study.J Biomol Struct Dyn. 2022 Jan;40(1):190-203. doi: 10.1080/07391102.2020.1810778. Epub 2020 Aug 27. J Biomol Struct Dyn. 2022. PMID: 32851919 Free PMC article.
-
Identification of Kaempferol as Viral Entry Inhibitor and DL-Arginine as Viral Replication Inhibitor from Selected Plants of Indian Traditional Medicine against COVID-19: An in silico Guided in vitro Approach.Curr Comput Aided Drug Des. 2023;19(4):313-323. doi: 10.2174/1573409919666230112123213. Curr Comput Aided Drug Des. 2023. PMID: 36635906
Cited by
-
Promising Role of Emodin as Therapeutics to Against Viral Infections.Front Pharmacol. 2022 May 4;13:902626. doi: 10.3389/fphar.2022.902626. eCollection 2022. Front Pharmacol. 2022. PMID: 35600857 Free PMC article. Review.
-
Screening of anti-Acinetobacter baumannii phytochemicals, based on the potential inhibitory effect on OmpA and OmpW functions.R Soc Open Sci. 2021 Aug 18;8(8):201652. doi: 10.1098/rsos.201652. eCollection 2021 Aug. R Soc Open Sci. 2021. PMID: 34457318 Free PMC article.
-
A comprehensive review on traditional applications, phytochemistry, pharmacology, and toxicology of Thymus serpyllum.Indian J Pharmacol. 2023 Nov-Dec;55(6):385-394. doi: 10.4103/ijp.ijp_220_22. Indian J Pharmacol. 2023. PMID: 38174535 Free PMC article. Review.
-
Methodology of network pharmacology for research on Chinese herbal medicine against COVID-19: A review.J Integr Med. 2022 Nov;20(6):477-487. doi: 10.1016/j.joim.2022.09.004. Epub 2022 Sep 22. J Integr Med. 2022. PMID: 36182651 Free PMC article. Review.
-
Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/ 3CLpro: molecular docking and simulation studies of three pertinent medicinal plant natural components.Curr Res Pharmacol Drug Discov. 2021;2:100038. doi: 10.1016/j.crphar.2021.100038. Epub 2021 Jun 5. Curr Res Pharmacol Drug Discov. 2021. PMID: 34870149 Free PMC article.
References
-
- Ahmed, S., & Shohael, A. M. (2019). In silico studies of four anthraquinones of Senna alataL. as potential antifungal compounds. Pharmacologyonline, 2, 259–268.
-
- Aja, P. M., Nwachukwu, N., Ibiam, U. A., Igwenyi, I. O., Offor, C. E., & Orji, U. O. (2014). Chemical constituents of Moringa oleifera leaves and seeds from Abakaliki, Nigeria. Am. J. Phytomed. Clin. Ther, 2, 310–321.
-
- Baric, R. S., Nelson, G. W., Fleming, J. O., Deans, R. J., Keck, J. G., Casteel, N., & Stohlman, S. A. (1988). Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. Journal of Virology, 62(11), 4280–4287. 10.1128/JVI.62.11.4280-4287.1988 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
