Contribution of plasma cells and B cells to hidradenitis suppurativa pathogenesis
- PMID: 32853177
- PMCID: PMC7566715
- DOI: 10.1172/jci.insight.139930
Contribution of plasma cells and B cells to hidradenitis suppurativa pathogenesis
Abstract
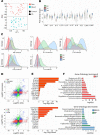
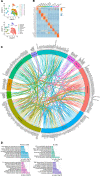
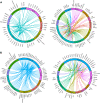
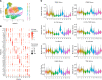
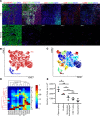
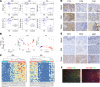
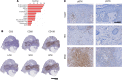
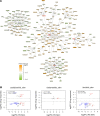
Hidradenitis suppurativa (HS) is a debilitating chronic inflammatory skin disease characterized by chronic abscess formation and development of multiple draining sinus tracts in the groin, axillae, and perineum. Using proteomic and transcriptomic approaches, we characterized the inflammatory responses in HS in depth, revealing immune responses centered on IFN-γ, IL-36, and TNF, with lesser contribution from IL-17A. We further identified B cells and plasma cells, with associated increases in immunoglobulin production and complement activation, as pivotal players in HS pathogenesis, with Bruton's tyrosine kinase (BTK) and spleen tyrosine kinase (SYK) pathway activation as a central signal transduction network in HS. These data provide preclinical evidence to accelerate the path toward clinical trials targeting BTK and SYK signaling in moderate-to-severe HS.
Keywords: B cells; Complement; Dermatology; Immunology; Skin.
Conflict of interest statement
Figures

References
-
- Jemec GB, Heidenheim M, Nielsen NH. The prevalence of hidradenitis suppurativa and its potential precursor lesions. J Am Acad Dermatol. 1996;35(2 pt 1):191–194. - PubMed
-
- Kohorst JJ, Kimball AB, Davis MD. Systemic associations of hidradenitis suppurativa. J Am Acad Dermatol. 2015;73(5 suppl 1):S27–S35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI139207/AI/NIAID NIH HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- S10 OD020053/OD/NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- R25 GM086262/GM/NIGMS NIH HHS/United States
- R01 AR075864/AR/NIAMS NIH HHS/United States
- P30 DK081943/DK/NIDDK NIH HHS/United States
- R01 AR069071/AR/NIAMS NIH HHS/United States
- R01 AI022553/AI/NIAID NIH HHS/United States
- R01 AR074302/AR/NIAMS NIH HHS/United States
- K08 AR060802/AR/NIAMS NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- P30 AR075043/AR/NIAMS NIH HHS/United States
- K01 AR072129/AR/NIAMS NIH HHS/United States
- R01 AR040312/AR/NIAMS NIH HHS/United States
- T32 AR007197/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

