Genomic Sequencing for Newborn Screening: Results of the NC NEXUS Project
- PMID: 32853555
- PMCID: PMC7536575
- DOI: 10.1016/j.ajhg.2020.08.001
Genomic Sequencing for Newborn Screening: Results of the NC NEXUS Project
Abstract
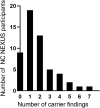
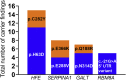
Newborn screening (NBS) was established as a public health program in the 1960s and is crucial for facilitating detection of certain medical conditions in which early intervention can prevent serious, life-threatening health problems. Genomic sequencing can potentially expand the screening for rare hereditary disorders, but many questions surround its possible use for this purpose. We examined the use of exome sequencing (ES) for NBS in the North Carolina Newborn Exome Sequencing for Universal Screening (NC NEXUS) project, comparing the yield from ES used in a screening versus a diagnostic context. We enrolled healthy newborns and children with metabolic diseases or hearing loss (106 participants total). ES confirmed the participant's underlying diagnosis in 15 out of 17 (88%) children with metabolic disorders and in 5 out of 28 (∼18%) children with hearing loss. We discovered actionable findings in four participants that would not have been detected by standard NBS. A subset of parents was eligible to receive additional information for their child about childhood-onset conditions with low or no clinical actionability, clinically actionable adult-onset conditions, and carrier status for autosomal-recessive conditions. We found pathogenic variants associated with hereditary breast and/or ovarian cancer in two children, a likely pathogenic variant in the gene associated with Lowe syndrome in one child, and an average of 1.8 reportable variants per child for carrier results. These results highlight the benefits and limitations of using genomic sequencing for NBS and the challenges of using such technology in future precision medicine approaches.
Keywords: exome sequencing; hearing loss; inborn errors of metabolism; newborn screening; pathogenic.
Copyright © 2020 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
D.B.B. is involved in an unrelated research study that receives contributed equipment and reagents from Asuragen and an unrelated research study that receives partial funding from Sarepta Therapeutics. B.C.P. is an investigator on a different research study that receives in-kind support (reagents and sequencing consumables) from Illumina (San Diego, CA). K.E.W. is President and Chair of the Executive Committee and Board of Directors for the Association for Molecular Pathology, a member of the Advisory Committee for the US FDA Medical Devices Molecular and Clinical Genetics Devices Panel, and a member of the Consultant Advisory Panel for BlueCross BlueShield of North Carolina.
Figures
References
-
- Guthrie R., Susi A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics. 1963;32:338–343. - PubMed
-
- Frazier D.M., Millington D.S., McCandless S.E., Koeberl D.D., Weavil S.D., Chaing S.H., Muenzer J. The tandem mass spectrometry newborn screening experience in North Carolina: 1997-2005. J. Inherit. Metab. Dis. 2006;29:76–85. - PubMed
-
- Centers for Disease Control and Prevention (CDC) Impact of expanded newborn screening--United States, 2006. MMWR Morb. Mortal. Wkly. Rep. 2008;57:1012–1015. - PubMed
-
- Hammett-Stabler C.A., Garg U. Humana Press; 2010. Clinical Applications of Mass Spectrometry Methods and Protocols. - PubMed
-
- Watson M.S., Mann M.Y., Lloyd-Puryear M.A., Rinaldo P., Howell R.R., Cordero J., Edwards E.S., Howse J.L., Mullaley T., Van Dyck P., American College of Medical Genetics Newborn Screening Expert Group Newborn screening: toward a uniform screening panel and system--executive summary. Pediatrics. 2006;117:S296–S307. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

