Structurally plastic NEMO and oligomerization prone IKK2 subunits define the behavior of human IKK2:NEMO complexes in solution
- PMID: 32853772
- PMCID: PMC7994000
- DOI: 10.1016/j.bbapap.2020.140526
Structurally plastic NEMO and oligomerization prone IKK2 subunits define the behavior of human IKK2:NEMO complexes in solution
Abstract
The human IκB Kinase (IKK) is a multisubunit protein complex of two kinases and one scaffolding subunit that controls induction of transcription factor NF-κB activity. IKK behaves as an entity of aberrantly high apparent molecular weight in solution. Recent X-ray crystallographic and cryo-electron microscopy structures of individual catalytic subunits (IKK1/IKKα and IKK2/IKKβ) reveal that they are both stably folded dimeric proteins that engage in extensive homo-oligomerization through unique surfaces that are required for activation of their respective catalytic activities. The NEMO/IKKγ subunit is a predominantly coiled coil protein that is required for activation of IKK through the canonical NF-κB signaling pathway. Here we report size-exclusion chromatography, multi-angle light scattering, analytical centrifugation, and thermal denaturation analyses of full-length human recombinant NEMO as well as deletion and disease-linked variants. We observe that NEMO is predominantly a dimer in solution, although by virtue of its modular coiled coil regions NEMO exhibits complicated solution dynamics involving portions that are mutually antagonistic toward homodimerization. This behavior causes NEMO to behave as a significantly larger sized particle in solution. Analyses of NEMO in complex with IKK2 indicate that NEMO preserves this structurally dynamic character within the multisubuit complex and provides the complex-bound IKK2 further propensity toward homo-oligomerization. These observations provide critical information on the structural plasticity of NEMO subunit dimers which helps clarify its role in diseases and in IKK regulation through oligomerization-dependent phosphorylation of catalytic IKK2 subunit dimers.
Keywords: Analytical ultracentrifugation; Circular dichroism spectroscopy; IκB kinase; Multi-angle light scattering; NEMO; NF-κB; Size-exclusion chromatography.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

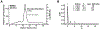

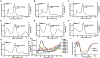
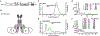
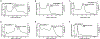

References
-
- Hayden MS, Ghosh S, Shared principles in NF-κB signaling, Cell 132 (2008) 344–362. - PubMed
-
- Mulero MC, Huxford T, Ghosh G, NF-κB, IκB, and IKK: integral components of immune system signaling, Adv. Exp. Med. Biol 1172 (2019) 207–226. - PubMed
-
- Scheidereit C, IκB kinase complexes: gateways to NF-κB activation and transcription, Oncogene 25 (2006) 6685–6705. - PubMed
-
- Chen ZJ, Parent L, Maniatis T, Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity, Cell 84 (1996) 853–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

