Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR
- PMID: 32853988
- PMCID: PMC7444471
- DOI: 10.1016/j.ebiom.2020.102960
Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR
Abstract
Background: Some COVID-19 cases test positive again for SARS-CoV-2 RNA following negative test results and discharge, raising questions about the meaning of virus detection. Better characterization of re-positive cases is urgently needed.
Methods: Clinical data were obtained through Guangdong's COVID-19 surveillance network. Neutralization antibody titre was determined using microneutralization assays. Potential infectivity of clinical samples was evaluated by cell inoculation. SARS-CoV-2 RNA was detected using three different RT-PCR kits and multiplex PCR with nanopore sequencing.
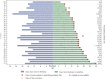
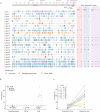
Findings: Among 619 discharged COVID-19 cases, 87 re-tested as SARS-CoV-2 positive in circumstances of social isolation. All re-positive cases had mild or moderate symptoms at initial diagnosis and were younger on average (median, 28). Re-positive cases (n = 59) exhibited similar neutralization antibodies (NAbs) titre distributions to other COVID-19 cases (n = 218) tested here. No infectious strain could be obtained by culture and no full-length viral genomes could be sequenced from re-positive cases.
Interpretation: Re-positive SARS-CoV-2 cases do not appear to be caused by active reinfection and were identified in ~14% of discharged cases. A robust NAb response and potential virus genome degradation were detected in almost all re-positive cases, suggesting a substantially lower transmission risk, especially through respiratory routes.
Keywords: COVID-19; Neutralizing antibody; Re-positive; SARS-CoV-2; Virology.
Copyright © 2020. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interests All authors: No reported conflicts of interest.
Figures

References
-
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A. Virological assessment of hospitalized patients with COVID-2019. Nature [Internet] 2020 http://www.nature.com/articles/s41586-020-2196-x Apr 1 [cited 2020 Apr 10]; Available from: - PubMed
-
- He X., Lau E.H.Y., Wu P., Deng X., Wang J., Hao X. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020:1–4. Apr 15. - PubMed
-
- WHO . 2020. Coronavirus disease (COVID-2019) situation reports [Internet]https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio... Available from:
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

