Evaluation of Microsatellite Typing, ITS Sequencing, AFLP Fingerprinting, MALDI-TOF MS, and Fourier-Transform Infrared Spectroscopy Analysis of Candida auris
- PMID: 32854308
- PMCID: PMC7576496
- DOI: 10.3390/jof6030146
Evaluation of Microsatellite Typing, ITS Sequencing, AFLP Fingerprinting, MALDI-TOF MS, and Fourier-Transform Infrared Spectroscopy Analysis of Candida auris
Abstract
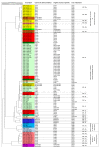
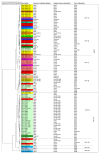
Candida auris is an emerging opportunistic yeast species causing nosocomial outbreaks at a global scale. A few studies have focused on the C. auris genotypic structure. Here, we compared five epidemiological typing tools using a set of 96 C. auris isolates from 14 geographical areas. Isolates were analyzed by microsatellite typing, ITS sequencing, amplified fragment length polymorphism (AFLP) fingerprint analysis, matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), and Fourier-transform infrared (FTIR) spectroscopy methods. Microsatellite typing grouped the isolates into four main clusters, corresponding to the four known clades in concordance with whole genome sequencing studies. The other investigated typing tools showed poor performance compared with microsatellite typing. A comparison between the five methods showed the highest agreement between microsatellite typing and ITS sequencing with 45% similarity, followed by microsatellite typing and the FTIR method with 33% similarity. The lowest agreement was observed between FTIR spectroscopy, MALDI-TOF MS, and ITS sequencing. This study indicates that microsatellite typing is the tool of choice for C. auris outbreak investigations. Additionally, FTIR spectroscopy requires further optimization and evaluation before it can be used as an epidemiological typing method, comparable with microsatellite typing, as a rapid method for tracing nosocomial fungal outbreaks.
Keywords: Candida auris; epidemiological typing; molecular epidemiology; nosocomial outbreak.
Conflict of interest statement
M.K., N.M., T.M., and M.V. are employees of the mass spectrometry company Bruker Daltonik GmbH. All authors declare that they have no competing interests. V.R. is an employee of the WI and CEO of BioAware and has no competing interest.
Figures



References
-
- Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., Colombo A.L., Calvo B., Cuomo C.A., Desjardins C.A., et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. - DOI - PMC - PubMed
-
- Schelenz S., Hagen F., Rhodes J.L., Abdolrasouli A., Chowdhary A., Hall A., Ryan L., Shackleton J., Trimlett R., Meis J.F., et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control. 2016;5:35. doi: 10.1186/s13756-016-0132-5. - DOI - PMC - PubMed
-
- Chowdhary A., Prakash A., Sharma C., Kordalewska M., Kumar A., Sarma S., Tarai B., Singh A., Upadhyaya G., Upadhyay S., et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-2017) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018;73:891–899. doi: 10.1093/jac/dkx480. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

