Exploring the Regulatory Potential of Long Non-Coding RNA in Feed Efficiency of Indicine Cattle
- PMID: 32854445
- PMCID: PMC7565090
- DOI: 10.3390/genes11090997
Exploring the Regulatory Potential of Long Non-Coding RNA in Feed Efficiency of Indicine Cattle
Abstract
Long non-coding RNA (lncRNA) can regulate several aspects of gene expression, being associated with complex phenotypes in humans and livestock species. In taurine beef cattle, recent evidence points to the involvement of lncRNA in feed efficiency (FE), a proxy for increased productivity and sustainability. Here, we hypothesized specific regulatory roles of lncRNA in FE of indicine cattle. Using RNA-Seq data from the liver, muscle, hypothalamus, pituitary gland and adrenal gland from Nellore bulls with divergent FE, we submitted new transcripts to a series of filters to confidently predict lncRNA. Then, we identified lncRNA that were differentially expressed (DE) and/or key regulators of FE. Finally, we explored lncRNA genomic location and interactions with miRNA and mRNA to infer potential function. We were able to identify 126 relevant lncRNA for FE in Bos indicus, some with high homology to previously identified lncRNA in Bos taurus and some possible specific regulators of FE in indicine cattle. Moreover, lncRNA identified here were linked to previously described mechanisms related to FE in hypothalamus-pituitary-adrenal axis and are expected to help elucidate this complex phenotype. This study contributes to expanding the catalogue of lncRNA, particularly in indicine cattle, and identifies candidates for further studies in animal selection and management.
Keywords: Bos indicus; RNA-Seq; co-expression network; residual feed intake.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
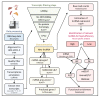
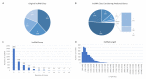
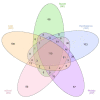
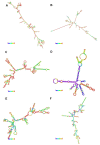

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

