Comprehensive analysis of prognostic tumor microenvironment-related genes in osteosarcoma patients
- PMID: 32854645
- PMCID: PMC7450807
- DOI: 10.1186/s12885-020-07216-2
Comprehensive analysis of prognostic tumor microenvironment-related genes in osteosarcoma patients
Abstract
Background: Tumor microenvironment (TME) plays an important role in malignant tumors. Our study aimed to investigate the effect of the TME and related genes in osteosarcoma patients.
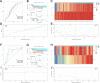
Methods: Gene expression profiles and clinical data of osteosarcoma patients were downloaded from the TARGET dataset. ESTIMATE algorithm was used to quantify the immune score. Then, the association between immune score and prognosis was studied. Afterward, a differential analysis was performed based on the high- and low-immune scores to determine TME-related genes. Additionally, Cox analyses were performed to construct two prognostic signatures for overall survival (OS) and disease-free survival (DFS), respectively. Two datasets obtained from the GEO database were used to validate signatures.
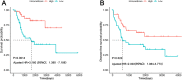
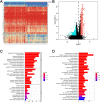
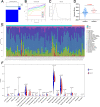
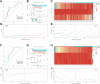
Results: Eighty-five patients were included in our research. The survival analysis indicated that patients with higher immune score have a favorable OS and DFS. Moreover, 769 genes were determined as TME-related genes. The unsupervised clustering analysis revealed two clusters were significantly related to immune score and T cells CD4 memory fraction. In addition, two signatures were generated based on three and two TME-related genes, respectively. Both two signatures can significantly divide patients into low- and high-risk groups and were validated in two GEO datasets. Afterward, the risk score and metastatic status were identified as independent prognostic factors for both OS and DFS and two nomograms were generated. The C-indexes of OS nomogram and DFS nomogram were 0.791 and 0.711, respectively.
Conclusion: TME was associated with the prognosis of osteosarcoma patients. Prognostic models based on TME-related genes can effectively predict OS and DFS of osteosarcoma patients.
Keywords: Immune features; Nomogram; Osteosarcoma; Prognosis; Tumor microenvironment.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Jaffe N, Bruland OS, Bielack S. Pediatric and adolescent osteosarcoma, vol. 152. New York: Springer Science & Business Media; 2010.
-
- Vander RG. Osteosarcoma and its variants. Orthopedic Clin North Am. 1996;27(3):575–581. - PubMed
-
- Biermann JS, Adkins D, Benjamin R, Brigman B, Chow W, Conrad EU, 3rd, Frassica D, Frassica FJ, George S, Healey JH, et al. Bone cancer. J Natl Compr Cancer Netw. 2007;5(4):420–437. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

