Clinically relevant radioresistant rhabdomyosarcoma cell lines: functional, molecular and immune-related characterization
- PMID: 32854690
- PMCID: PMC7453562
- DOI: 10.1186/s12929-020-00683-6
Clinically relevant radioresistant rhabdomyosarcoma cell lines: functional, molecular and immune-related characterization
Abstract
Background: The probability of local tumor control after radiotherapy (RT) remains still miserably poor in pediatric rhabdomyosarcoma (RMS). Thus, understanding the molecular mechanisms responsible of tumor relapse is essential to identify personalized RT-based strategies. Contrary to what has been done so far, a correct characterization of cellular radioresistance should be performed comparing radioresistant and radiosensitive cells with the same isogenic background.
Methods: Clinically relevant radioresistant (RR) embryonal (RD) and alveolar (RH30) RMS cell lines have been developed by irradiating them with clinical-like hypo-fractionated schedule. RMS-RR cells were compared to parental isogenic counterpart (RMS-PR) and studied following the radiobiological concept of the "6Rs", which stand for repair, redistribution, repopulation, reoxygenation, intrinsic radioresistance and radio-immuno-biology.
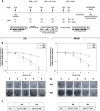
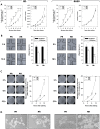
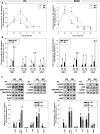
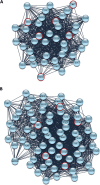
Results: RMS-RR cell lines, characterized by a more aggressive and in vitro pro-metastatic phenotype, showed a higher ability to i) detoxify from reactive oxygen species; ii) repair DNA damage by differently activating non-homologous end joining and homologous recombination pathways; iii) counteract RT-induced G2/M cell cycle arrest by re-starting growth and repopulating after irradiation; iv) express cancer stem-like profile. Bioinformatic analyses, performed to assess the role of 41 cytokines after RT exposure and their network interactions, suggested TGF-β, MIF, CCL2, CXCL5, CXCL8 and CXCL12 as master regulators of cancer immune escape in RMS tumors.
Conclusions: These results suggest that RMS could sustain intrinsic and acquire radioresistance by different mechanisms and indicate potential targets for future combined radiosensitizing strategies.
Keywords: Immunoescape; Radiobiology; Radioresistance; Radiotherapy; Rhabdomyosarcoma.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- O'Sullivan B, Davis AM, Turcotte R, Bell R, Catton C, Chabot P, Wunder J, Kandel R, Goddard K, Sadura A, Pater J, Zee B. Preoperative versus postoperative radiotherapy in soft-tissue sarcoma of the limbs: a randomised trial. Lancet. 2002;359:2235–2241. - PubMed
-
- Golden EB, Apetoh L. Radiotherapy and immunogenic cell death. Semin Radiat Oncol. 2015;25:11–17. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

