The dynamic landscape of transcription initiation in yeast mitochondria
- PMID: 32855416
- PMCID: PMC7452894
- DOI: 10.1038/s41467-020-17793-2
The dynamic landscape of transcription initiation in yeast mitochondria
Abstract
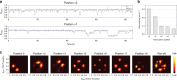
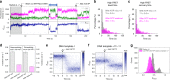
Controlling efficiency and fidelity in the early stage of mitochondrial DNA transcription is crucial for regulating cellular energy metabolism. Conformational transitions of the transcription initiation complex must be central for such control, but how the conformational dynamics progress throughout transcription initiation remains unknown. Here, we use single-molecule fluorescence resonance energy transfer techniques to examine the conformational dynamics of the transcriptional system of yeast mitochondria with single-base resolution. We show that the yeast mitochondrial transcriptional complex dynamically transitions among closed, open, and scrunched states throughout the initiation stage. Then abruptly at position +8, the dynamic states of initiation make a sharp irreversible transition to an unbent conformation with associated promoter release. Remarkably, stalled initiation complexes remain in dynamic scrunching and unscrunching states without dissociating the RNA transcript, implying the existence of backtracking transitions with possible regulatory roles. The dynamic landscape of transcription initiation suggests a kinetically driven regulation of mitochondrial transcription.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Cryo-EM Structures Reveal Transcription Initiation Steps by Yeast Mitochondrial RNA Polymerase.Mol Cell. 2021 Jan 21;81(2):268-280.e5. doi: 10.1016/j.molcel.2020.11.016. Epub 2020 Dec 4. Mol Cell. 2021. PMID: 33278362 Free PMC article.
-
The C-terminal tail of the yeast mitochondrial transcription factor Mtf1 coordinates template strand alignment, DNA scrunching and timely transition into elongation.Nucleic Acids Res. 2020 Mar 18;48(5):2604-2620. doi: 10.1093/nar/gkaa040. Nucleic Acids Res. 2020. PMID: 31980825 Free PMC article.
-
Fluorescence mapping of the open complex of yeast mitochondrial RNA polymerase.J Biol Chem. 2009 Feb 27;284(9):5514-22. doi: 10.1074/jbc.M807880200. Epub 2008 Dec 30. J Biol Chem. 2009. PMID: 19116203 Free PMC article.
-
Mechanism of transcription initiation by the yeast mitochondrial RNA polymerase.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):930-8. doi: 10.1016/j.bbagrm.2012.02.003. Epub 2012 Feb 14. Biochim Biophys Acta. 2012. PMID: 22353467 Free PMC article. Review.
-
The impact of transcription on posttranscriptional processes in yeast.Gene. 2013 Aug 15;526(1):23-9. doi: 10.1016/j.gene.2013.04.021. Epub 2013 Apr 30. Gene. 2013. PMID: 23639960 Review.
Cited by
-
Cryo-EM Structures Reveal Transcription Initiation Steps by Yeast Mitochondrial RNA Polymerase.Mol Cell. 2021 Jan 21;81(2):268-280.e5. doi: 10.1016/j.molcel.2020.11.016. Epub 2020 Dec 4. Mol Cell. 2021. PMID: 33278362 Free PMC article.
-
Structure, mechanism, and regulation of mitochondrial DNA transcription initiation.J Biol Chem. 2020 Dec 25;295(52):18406-18425. doi: 10.1074/jbc.REV120.011202. Epub 2020 Oct 30. J Biol Chem. 2020. PMID: 33127643 Free PMC article. Review.
-
Structures illustrate step-by-step mitochondrial transcription initiation.Nature. 2023 Oct;622(7984):872-879. doi: 10.1038/s41586-023-06643-y. Epub 2023 Oct 11. Nature. 2023. PMID: 37821701 Free PMC article.
-
The C-terminal tail of the yeast mitochondrial transcription factor Mtf1 coordinates template strand alignment, DNA scrunching and timely transition into elongation.Nucleic Acids Res. 2020 Mar 18;48(5):2604-2620. doi: 10.1093/nar/gkaa040. Nucleic Acids Res. 2020. PMID: 31980825 Free PMC article.
-
Real-Time Single-Molecule Studies of RNA Polymerase-Promoter Open Complex Formation Reveal Substantial Heterogeneity Along the Promoter-Opening Pathway.J Mol Biol. 2022 Jan 30;434(2):167383. doi: 10.1016/j.jmb.2021.167383. Epub 2021 Dec 1. J Mol Biol. 2022. PMID: 34863780 Free PMC article.
References
-
- Gralla JD, Carpousis AJ, Stefano JE. Productive and abortive initiation of transcription in vitro at the lac UV5 promoter. Biochemistry. 1980;19:5864–5869. - PubMed
-
- Kubori T, Shimamoto N. A branched pathway in the early stage of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 1996;256:449–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

