Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion
- PMID: 32855419
- PMCID: PMC7452896
- DOI: 10.1038/s41467-020-18066-8
Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion
Abstract
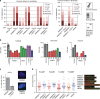
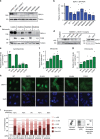
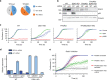
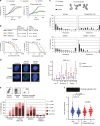
Warsaw Breakage Syndrome (WABS) is a rare disorder related to cohesinopathies and Fanconi anemia, caused by bi-allelic mutations in DDX11. Here, we report multiple compound heterozygous WABS cases, each displaying destabilized DDX11 protein and residual DDX11 function at the cellular level. Patient-derived cell lines exhibit sensitivity to topoisomerase and PARP inhibitors, defective sister chromatid cohesion and reduced DNA replication fork speed. Deleting DDX11 in RPE1-TERT cells inhibits proliferation and survival in a TP53-dependent manner and causes chromosome breaks and cohesion defects, independent of the expressed pseudogene DDX12p. Importantly, G-quadruplex (G4) stabilizing compounds induce chromosome breaks and cohesion defects which are strongly aggravated by inactivation of DDX11 but not FANCJ. The DNA helicase domain of DDX11 is essential for sister chromatid cohesion and resistance to G4 stabilizers. We propose that DDX11 is a DNA helicase protecting against G4 induced double-stranded breaks and concomitant loss of cohesion, possibly at DNA replication forks.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bailey C, Fryer AE, Greenslade M. Warsaw breakage syndrome—a further report, emphasising cutaneous findings. Eur. J. Med. Genet. 2015;58:235–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

