Evaluation of SARS-CoV-2 serology assays reveals a range of test performance
- PMID: 32855547
- PMCID: PMC7740072
- DOI: 10.1038/s41587-020-0659-0
Evaluation of SARS-CoV-2 serology assays reveals a range of test performance
Abstract
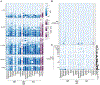
Appropriate use and interpretation of serological tests for assessments of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) exposure, infection and potential immunity require accurate data on assay performance. We conducted a head-to-head evaluation of ten point-of-care-style lateral flow assays (LFAs) and two laboratory-based enzyme-linked immunosorbent assays to detect anti-SARS-CoV-2 IgM and IgG antibodies in 5-d time intervals from symptom onset and studied the specificity of each assay in pre-coronavirus disease 2019 specimens. The percent of seropositive individuals increased with time, peaking in the latest time interval tested (>20 d after symptom onset). Test specificity ranged from 84.3% to 100.0% and was predominantly affected by variability in IgM results. LFA specificity could be increased by considering weak bands as negative, but this decreased detection of antibodies (sensitivity) in a subset of SARS-CoV-2 real-time PCR-positive cases. Our results underline the importance of seropositivity threshold determination and reader training for reliable LFA deployment. Although there was no standout serological assay, four tests achieved more than 80% positivity at later time points tested and more than 95% specificity.
Figures
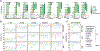
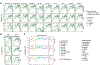
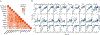
Update of
-
Test performance evaluation of SARS-CoV-2 serological assays.medRxiv [Preprint]. 2020 May 17:2020.04.25.20074856. doi: 10.1101/2020.04.25.20074856. medRxiv. 2020. Update in: Nat Biotechnol. 2020 Oct;38(10):1174-1183. doi: 10.1038/s41587-020-0659-0. PMID: 32511497 Free PMC article. Updated. Preprint.
References
-
- Johns Hopkins University. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU), <https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594...> (2020).
-
- Infectious Diseases Society of America. IDSA Statement on COVID-19 Testing, <https://www.idsociety.org/globalassets/idsa/public-health/covid-19-idsa-...> (2020).
-
- World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. (World Health Organization, Geneva, 2020).
Publication types
MeSH terms
Substances
Grants and funding
- R38 HL143581/HL/NHLBI NIH HHS/United States
- F30AI150061/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- T32GM007618/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- 1F31NS106868-01/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- U01CK000490/ACL/ACL HHS/United States
- T32 CA108462/CA/NCI NIH HHS/United States
- F31 NS106868/NS/NINDS NIH HHS/United States
- F30 AI147364/AI/NIAID NIH HHS/United States
- R01 AI040098/AI/NIAID NIH HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- R38HL143581/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- T32 GM007618/GM/NIGMS NIH HHS/United States
- 1F30HD093116/U.S. Department of Health & Human Services | National Institutes of Health (NIH)/International
- U19 AI135990/AI/NIAID NIH HHS/United States
- L40 AI140341/AI/NIAID NIH HHS/United States
- F30 AI150061/AI/NIAID NIH HHS/United States
- U01 CK000490/CK/NCEZID CDC HHS/United States
- K38 HL154203/HL/NHLBI NIH HHS/United States
- F30 HD093116/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

