Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species
- PMID: 32855739
- PMCID: PMC7426331
- DOI: 10.1016/j.simyco.2020.05.002
Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species
Abstract
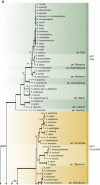
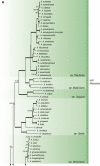
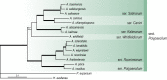
The Eurotiales is a relatively large order of Ascomycetes with members frequently having positive and negative impact on human activities. Species within this order gain attention from various research fields such as food, indoor and medical mycology and biotechnology. In this article we give an overview of families and genera present in the Eurotiales and introduce an updated subgeneric, sectional and series classification for Aspergillus and Penicillium. Finally, a comprehensive list of accepted species in the Eurotiales is given. The classification of the Eurotiales at family and genus level is traditionally based on phenotypic characters, and this classification has since been challenged using sequence-based approaches. Here, we re-evaluated the relationships between families and genera of the Eurotiales using a nine-gene sequence dataset. Based on this analysis, the new family Penicillaginaceae is introduced and four known families are accepted: Aspergillaceae, Elaphomycetaceae, Thermoascaceae and Trichocomaceae. The Eurotiales includes 28 genera: 15 genera are accommodated in the Aspergillaceae (Aspergillago, Aspergillus, Evansstolkia, Hamigera, Leiothecium, Monascus, Penicilliopsis, Penicillium, Phialomyces, Pseudohamigera, Pseudopenicillium, Sclerocleista, Warcupiella, Xerochrysium and Xeromyces), eight in the Trichocomaceae (Acidotalaromyces, Ascospirella, Dendrosphaera, Rasamsonia, Sagenomella, Talaromyces, Thermomyces, Trichocoma), two in the Thermoascaceae (Paecilomyces, Thermoascus) and one in the Penicillaginaceae (Penicillago). The classification of the Elaphomycetaceae was not part of this study, but according to literature two genera are present in this family (Elaphomyces and Pseudotulostoma). The use of an infrageneric classification system has a long tradition in Aspergillus and Penicillium. Most recent taxonomic studies focused on the sectional level, resulting in a well-established sectional classification in these genera. In contrast, a series classification in Aspergillus and Penicillium is often outdated or lacking, but is still relevant, e.g., the allocation of a species to a series can be highly predictive in what functional characters the species might have and might be useful when using a phenotype-based identification. The majority of the series in Aspergillus and Penicillium are invalidly described and here we introduce a new series classification. Using a phylogenetic approach, often supported by phenotypic, physiologic and/or extrolite data, Aspergillus is subdivided in six subgenera, 27 sections (five new) and 75 series (73 new, one new combination), and Penicillium in two subgenera, 32 sections (seven new) and 89 series (57 new, six new combinations). Correct identification of species belonging to the Eurotiales is difficult, but crucial, as the species name is the linking pin to information. Lists of accepted species are a helpful aid for researchers to obtain a correct identification using the current taxonomic schemes. In the most recent list from 2014, 339 Aspergillus, 354 Penicillium and 88 Talaromyces species were accepted. These numbers increased significantly, and the current list includes 446 Aspergillus (32 % increase), 483 Penicillium (36 % increase) and 171 Talaromyces (94 % increase) species, showing the large diversity and high interest in these genera. We expanded this list with all genera and species belonging to the Eurotiales (except those belonging to Elaphomycetaceae). The list includes 1 187 species, distributed over 27 genera, and contains MycoBank numbers, collection numbers of type and ex-type cultures, subgenus, section and series classification data, information on the mode of reproduction, and GenBank accession numbers of ITS, beta-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) gene sequences.
Keywords: Acidotalaromyces Houbraken, Frisvad & Samson; Acidotalaromyces lignorum (Stolk) Houbraken, Frisvad & Samson; Ascospirella Houbraken, Frisvad & Samson; Ascospirella lutea (Zukal) Houbraken, Frisvad & Samson; Aspergillus chaetosartoryae Hubka, Kocsubé & Houbraken; Classification; Evansstolkia Houbraken, Frisvad & Samson; Evansstolkia leycettana (H.C. Evans & Stolk) Houbraken, Frisvad & Samson; Hamigera brevicompacta (H.Z. Kong) Houbraken, Frisvad & Samson; Infrageneric classification; New combinations, series; New combinations, species; New genera; New names; New sections; New series; New taxa; Nomenclature; Paecilomyces lagunculariae (C. Ram) Houbraken, Frisvad & Samson; Penicillaginaceae Houbraken, Frisvad & Samson; Penicillago kabunica (Baghd.) Houbraken, Frisvad & Samson; Penicillago mirabilis (Beliakova & Milko) Houbraken, Frisvad & Samson; Penicillago moldavica (Milko & Beliakova) Houbraken, Frisvad & Samson; Phialomyces arenicola (Chalab.) Houbraken, Frisvad & Samson; Phialomyces humicoloides (Bills & Heredia) Houbraken, Frisvad & Samson; Phylogeny; Polythetic classes; Pseudohamigera Houbraken, Frisvad & Samson; Pseudohamigera striata (Raper & Fennell) Houbraken, Frisvad & Samson; Talaromyces resinae (Z.T. Qi & H.Z. Kong) Houbraken & X.C. Wang; Talaromyces striatoconidius Houbraken, Frisvad & Samson; Taxonomic novelties: New family; Thermoascus verrucosus (Samson & Tansey) Houbraken, Frisvad & Samson; Thermoascus yaguchii Houbraken, Frisvad & Samson; in Aspergillus: sect. Bispori S.W. Peterson, Varga, Frisvad, Samson ex Houbraken; in Aspergillus: ser. Acidohumorum Houbraken & Frisvad; in Aspergillus: ser. Inflati (Stolk & Samson) Houbraken & Frisvad; in Penicillium: sect. Alfrediorum Houbraken & Frisvad; in Penicillium: ser. Adametziorum Houbraken & Frisvad; in Penicillium: ser. Alutacea (Pitt) Houbraken & Frisvad; sect. Crypta Houbraken & Frisvad; sect. Eremophila Houbraken & Frisvad; sect. Formosana Houbraken & Frisvad; sect. Griseola Houbraken & Frisvad; sect. Inusitata Houbraken & Frisvad; sect. Lasseniorum Houbraken & Frisvad; sect. Polypaecilum Houbraken & Frisvad; sect. Raperorum S.W. Peterson, Varga, Frisvad, Samson ex Houbraken; sect. Silvatici S.W. Peterson, Varga, Frisvad, Samson ex Houbraken; sect. Vargarum Houbraken & Frisvad; ser. Alliacei Houbraken & Frisvad; ser. Ambigui Houbraken & Frisvad; ser. Angustiporcata Houbraken & Frisvad; ser. Arxiorum Houbraken & Frisvad; ser. Atramentosa Houbraken & Frisvad; ser. Aurantiobrunnei Houbraken & Frisvad; ser. Avenacei Houbraken & Frisvad; ser. Bertholletiarum Houbraken & Frisvad; ser. Biplani Houbraken & Frisvad; ser. Brevicompacta Houbraken & Frisvad; ser. Brevipedes Houbraken & Frisvad; ser. Brunneouniseriati Houbraken & Frisvad; ser. Buchwaldiorum Houbraken & Frisvad; ser. Calidousti Houbraken & Frisvad; ser. Canini Houbraken & Frisvad; ser. Carbonarii Houbraken & Frisvad; ser. Cavernicolarum Houbraken & Frisvad; ser. Cervini Houbraken & Frisvad; ser. Chevalierorum Houbraken & Frisvad; ser. Cinnamopurpurea Houbraken & Frisvad; ser. Circumdati Houbraken & Frisvad; ser. Clavigera Houbraken & Frisvad; ser. Conjuncti Houbraken & Frisvad; ser. Copticolarum Houbraken & Frisvad; ser. Coremiiformes Houbraken & Frisvad; ser. Corylophila Houbraken & Frisvad; ser. Costaricensia Houbraken & Frisvad; ser. Cremei Houbraken & Frisvad; ser. Crustacea (Pitt) Houbraken & Frisvad; ser. Dalearum Houbraken & Frisvad; ser. Deflecti Houbraken & Frisvad; ser. Egyptiaci Houbraken & Frisvad; ser. Erubescentia (Pitt) Houbraken & Frisvad; ser. Estinogena Houbraken & Frisvad; ser. Euglauca Houbraken & Frisvad; ser. Fennelliarum Houbraken & Frisvad; ser. Flavi Houbraken & Frisvad; ser. Flavipedes Houbraken & Frisvad; ser. Fortuita Houbraken & Frisvad; ser. Fumigati Houbraken & Frisvad; ser. Funiculosi Houbraken & Frisvad; ser. Gallaica Houbraken & Frisvad; ser. Georgiensia Houbraken & Frisvad; ser. Goetziorum Houbraken & Frisvad; ser. Gracilenta Houbraken & Frisvad; ser. Halophilici Houbraken & Frisvad; ser. Herqueorum Houbraken & Frisvad; ser. Heteromorphi Houbraken & Frisvad; ser. Hoeksiorum Houbraken & Frisvad; ser. Homomorphi Houbraken & Frisvad; ser. Idahoensia Houbraken & Frisvad; ser. Implicati Houbraken & Frisvad; ser. Improvisa Houbraken & Frisvad; ser. Indica Houbraken & Frisvad; ser. Japonici Houbraken & Frisvad; ser. Jiangxiensia Houbraken & Frisvad; ser. Kalimarum Houbraken & Frisvad; ser. Kiamaensia Houbraken & Frisvad; ser. Kitamyces Houbraken & Frisvad; ser. Lapidosa (Pitt) Houbraken & Frisvad; ser. Leporum Houbraken & Frisvad; ser. Leucocarpi Houbraken & Frisvad; ser. Livida Houbraken & Frisvad; ser. Longicatenata Houbraken & Frisvad; ser. Macrosclerotiorum Houbraken & Frisvad; ser. Monodiorum Houbraken & Frisvad; ser. Multicolores Houbraken & Frisvad; ser. Neoglabri Houbraken & Frisvad; ser. Neonivei Houbraken & Frisvad; ser. Nidulantes Houbraken & Frisvad; ser. Nigri Houbraken & Frisvad; ser. Nivei Houbraken & Frisvad; ser. Nodula Houbraken & Frisvad; ser. Nomiarum Houbraken & Frisvad; ser. Noonimiarum Houbraken & Frisvad; ser. Ochraceorosei Houbraken & Frisvad; ser. Olivimuriarum Houbraken & Frisvad; ser. Osmophila Houbraken & Frisvad; ser. Paradoxa Houbraken & Frisvad; ser. Paxillorum Houbraken & Frisvad; ser. Penicillioides Houbraken & Frisvad; ser. Phoenicea Houbraken & Frisvad; ser. Pinetorum (Pitt) Houbraken & Frisvad; ser. Polypaecilum Houbraken & Frisvad; ser. Pulvini Houbraken & Frisvad; ser. Quercetorum Houbraken & Frisvad; ser. Raistrickiorum Houbraken & Frisvad; ser. Ramigena Houbraken & Frisvad; ser. Restricti Houbraken & Frisvad; ser. Robsamsonia Houbraken & Frisvad; ser. Rolfsiorum Houbraken & Frisvad; ser. Roseopurpurea Houbraken & Frisvad; ser. Rubri Houbraken & Frisvad; ser. Salinarum Houbraken & Frisvad; ser. Samsoniorum Houbraken & Frisvad; ser. Saturniformia Houbraken & Frisvad; ser. Scabrosa Houbraken & Frisvad; ser. Sclerotigena Houbraken & Frisvad; ser. Sclerotiorum Houbraken & Frisvad; ser. Sheariorum Houbraken & Frisvad; ser. Simplicissima Houbraken & Frisvad; ser. Soppiorum Houbraken & Frisvad; ser. Sparsi Houbraken & Frisvad; ser. Spathulati Houbraken & Frisvad; ser. Spelaei Houbraken & Frisvad; ser. Speluncei Houbraken & Frisvad; ser. Spinulosa Houbraken & Frisvad; ser. Stellati Houbraken & Frisvad; ser. Steyniorum Houbraken & Frisvad; ser. Sublectatica Houbraken & Frisvad; ser. Sumatraensia Houbraken & Frisvad; ser. Tamarindosolorum Houbraken & Frisvad; ser. Teporium Houbraken & Frisvad; ser. Terrei Houbraken & Frisvad; ser. Thermomutati Houbraken & Frisvad; ser. Thiersiorum Houbraken & Frisvad; ser. Thomiorum Houbraken & Frisvad; ser. Unguium Houbraken & Frisvad; ser. Unilaterales Houbraken & Frisvad; ser. Usti Houbraken & Frisvad; ser. Verhageniorum Houbraken & Frisvad; ser. Versicolores Houbraken & Frisvad; ser. Virgata Houbraken & Frisvad; ser. Viridinutantes Houbraken & Frisvad; ser. Vitricolarum Houbraken & Frisvad; ser. Wentiorum Houbraken & Frisvad; ser. Westlingiorum Houbraken & Frisvad; ser. Whitfieldiorum Houbraken & Frisvad; ser. Xerophili Houbraken & Frisvad; series Tularensia (Pitt) Houbraken & Frisvad.
© 2020 Westerdijk Fungal Biodiversity Institute. Production and hosting by ELSEVIER B.V.
Figures


























References
-
- Afiyatullov S.S., Leshchenko E.V., Antonov A.S., et al. Secondary metabolites of fungus Penicillium thomii associated with eelgrass Zostera marina. Chemistry of Natural Compounds. 2018;54:1029–1030.
-
- Afiyatullov S.S., Leshchenko E.V., Berdyshev D.V., et al. Zosteropenillines: Polyketides from the marine-derived fungus Penicillium thomii. Marine Drugs. 2017;15:46.
-
- Afiyatullov S.S., Leshchenko E.V., Sobolevskaya M.P., et al. New thomimarine E from marine isolate of the fungus Penicillium thomii. Chemistry of Natural Compounds. 2017;53:290–294.
-
- Agurell S.L. Costaclavine from Penicillium chermesinum. Experientia. 1964;20:25–26. - PubMed
-
- Ahmad A., Akram W., Shahzadi I., et al. Benzenedicarboxylic acid upregulates O48814 and Q9FJQ8 for improved nutritional contents of tomato and low risk of fungal attack. Journal of the Science of Food and Agriculture. 2019;99:6139–6154. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
