Translesion synthesis by AMV, HIV, and MMLVreverse transcriptases using RNA templates containing inosine, guanosine, and their 8-oxo-7,8-dihydropurine derivatives
- PMID: 32857764
- PMCID: PMC7455023
- DOI: 10.1371/journal.pone.0235102
Translesion synthesis by AMV, HIV, and MMLVreverse transcriptases using RNA templates containing inosine, guanosine, and their 8-oxo-7,8-dihydropurine derivatives
Abstract
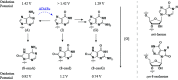
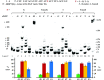
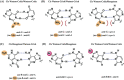
Inosine is ubiquitous and essential in many biological processes, including RNA-editing. In addition, oxidative stress on RNA has been a topic of increasing interest due, in part, to its potential role in the development/progression of disease. In this work we probed the ability of three reverse transcriptases (RTs) to catalyze the synthesis of cDNA in the presence of RNA templates containing inosine (I), 8-oxo-7,8-dihydroinosine (8oxo-I), guanosine (G), or 8-oxo-7,8-dihydroguanosine (8-oxoG), and explored the impact that these purine derivatives have as a function of position. To this end, we used 29-mers of RNA (as template) containing the modifications at position-18 and reverse transcribed DNA using 17-mers, 18-mers, or 19-mers (as primers). Generally reactivity of the viral RTs, AMV / HIV / MMLV, towards cDNA synthesis was similar for templates containing G or I as well as for those with 8-oxoG or 8-oxoI. Notable differences are: 1) the use of 18-mers of DNA (to explore cDNA synthesis past the lesion/modification) led to inhibition of DNA elongation in cases where a G:dA wobble pair was present, while the presence of I, 8-oxoI, or 8-oxoG led to full synthesis of the corresponding cDNA, with the latter two displaying a more efficient process; 2) HIV RT is more sensitive to modified base pairs in the vicinity of cDNA synthesis; and 3) the presence of a modification two positions away from transcription initiation has an adverse impact on the overall process. Steady-state kinetics were established using AMV RT to determine substrate specificities towards canonical dNTPs (N = G, C, T, A). Overall we found evidence that RNA templates containing inosine are likely to incorporate dC > dT > > dA, where reactivity in the presence of dA was found to be pH dependent (process abolished at pH 7.3); and that the absence of the C2-exocyclic amine, as displayed with templates containing 8-oxoI, leads to increased selectivity towards incorporation of dA over dC. The data will be useful in assessing the impact that the presence of inosine and/or oxidatively generated lesions have on viral processes and adds to previous reports where I codes exclusively like G. Similar results were obtained upon comparison of AMV and MMLV RTs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

