Analyzing and validating the prognostic value and mechanism of colon cancer immune microenvironment
- PMID: 32859214
- PMCID: PMC7456375
- DOI: 10.1186/s12967-020-02491-w
Analyzing and validating the prognostic value and mechanism of colon cancer immune microenvironment
Abstract
Background: Colon cancer is a disease with high malignancy and incidence in the world. Tumor immune microenvironment (TIM) and tumor mutational burden (TMB) have been proved to play crucial roles in predicting clinical outcomes and therapeutic efficacy, but the correlation between them and the underlying mechanism were not completely understood in colon cancer.
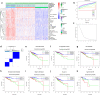
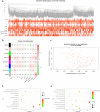
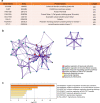
Methods: In this study, we used Single-Sample Gene Set Enrichment Analysis (ssGSEA) and unsupervised consensus clustering analysis to divide patients from the TCGA cohort into three immune subgroups. Then we validated their differences in immune cell infiltration, overall survival outcomes, clinical phenotypes and expression levels of HLA and checkpoint genes by Mann-Whitney tests. We performed weighted correlation network analysis (WGCNA) to obtain immunity-related module and hub genes. Then we explored the underlying mechanism of hub genes by gene set enrichment analysis (GSEA) and gene set evaluation analysis (GSVA). Finally, we gave an overall view of gene variants and verified the correlation between TIM and TMB by comparing microsatellite instability (MSI) and gene mutations among three immune subgroups.
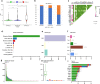
Results: The colon cancer patients were clustered into low immunity, median immunity and high immunity groups. The median immunity group had a favorable survival probability compared with that of the low and high immunity groups. Three groups had significant differences in immune cell infiltration, tumor stage, living state and T classification. We got 8 hub genes (CCDC69, CLMP, FAM110B, FAM129A, GUCY1B3, PALLD, PLEKHO1 and STY11) and predicted that immunity may correlated with inflammatory response, KRAS signaling pathway and T cell infiltration. With higher immunity, the TMB was higher. The most frequent mutations in low and median immunity groups were APC, TP53 and KRAS, while TTN and MUC16 showed higher mutational frequency in high immunity group.
Conclusions: We performed a comprehensive evaluation of the immune microenvironment landscape of colon cancer and demonstrated the positive correlation between immunity and TMB. The hub genes and frequently mutated genes were strongly related to immunity and may give suggestion for immunotherapy in the future.
Keywords: Colon cancer; Tumor immune microenvironment; Tumor mutational burden; Weighted correlation network analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

