Complete chemical structures of human mitochondrial tRNAs
- PMID: 32859890
- PMCID: PMC7455718
- DOI: 10.1038/s41467-020-18068-6
Complete chemical structures of human mitochondrial tRNAs
Abstract
Mitochondria generate most cellular energy via oxidative phosphorylation. Twenty-two species of mitochondrial (mt-)tRNAs encoded in mtDNA translate essential subunits of the respiratory chain complexes. mt-tRNAs contain post-transcriptional modifications introduced by nuclear-encoded tRNA-modifying enzymes. They are required for deciphering genetic code accurately, as well as stabilizing tRNA. Loss of tRNA modifications frequently results in severe pathological consequences. Here, we perform a comprehensive analysis of post-transcriptional modifications of all human mt-tRNAs, including 14 previously-uncharacterized species. In total, we find 18 kinds of RNA modifications at 137 positions (8.7% in 1575 nucleobases) in 22 species of human mt-tRNAs. An up-to-date list of 34 genes responsible for mt-tRNA modifications are provided. We identify two genes required for queuosine (Q) formation in mt-tRNAs. Our results provide insight into the molecular mechanisms underlying the decoding system and could help to elucidate the molecular pathogenesis of human mitochondrial diseases caused by aberrant tRNA modifications.
Conflict of interest statement
The authors declare no competing interests.
Figures



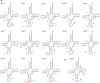

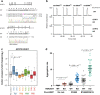
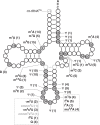
References
-
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Hallberg BM, Larsson NG. Making proteins in the powerhouse. Cell Metab. 2014;20:226–240. - PubMed
-
- Greber BJ, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

