Oncogenic AURKA-enhanced N6-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells
- PMID: 32859993
- PMCID: PMC8027457
- DOI: 10.1038/s41422-020-00397-2
Oncogenic AURKA-enhanced N6-methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem-like cells
Abstract
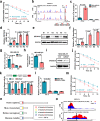
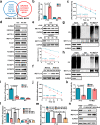
RNase III DROSHA is upregulated in multiple cancers and contributes to tumor progression by hitherto unclear mechanisms. Here, we demonstrate that DROSHA interacts with β-Catenin to transactivate STC1 in an RNA cleavage-independent manner, contributing to breast cancer stem-like cell (BCSC) properties. DROSHA mRNA stability is enhanced by N6-methyladenosine (m6A) modification which is activated by AURKA in BCSCs. AURKA stabilizes METTL14 by inhibiting its ubiquitylation and degradation to promote DROSHA mRNA methylation. Moreover, binding of AURKA to DROSHA transcript further strengthens the binding of the m6A reader IGF2BP2 to stabilize m6A-modified DROSHA. In addition, wild-type DROSHA, but not an m6A methylation-deficient mutant, enhances BCSC stemness maintenance, while inhibition of DROSHA m6A modification attenuates BCSC traits. Our study unveils the AURKA-induced oncogenic m6A modification as a key regulator of DROSHA in breast cancer and identifies a novel DROSHA transcriptional function in promoting the BCSC phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

