Detection of SARS-CoV-2 in raw and treated wastewater in Germany - Suitability for COVID-19 surveillance and potential transmission risks
- PMID: 32861187
- PMCID: PMC7434407
- DOI: 10.1016/j.scitotenv.2020.141750
Detection of SARS-CoV-2 in raw and treated wastewater in Germany - Suitability for COVID-19 surveillance and potential transmission risks
Abstract
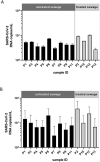
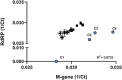
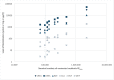
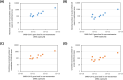
Wastewater-based monitoring of the spread of the new SARS-CoV-2 virus, also referred to as wastewater-based epidemiology (WBE), has been suggested as a tool to support epidemiology. An extensive sampling campaign, including nine municipal wastewater treatment plants, has been conducted in different cities of the Federal State of North Rhine-Westphalia (Germany) on the same day in April 2020, close to the first peak of the corona crisis. Samples were processed and analysed for a set of SARS-CoV-2-specific genes, as well as pan-genotypic gene sequences also covering other coronavirus types, using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Additionally, a comprehensive set of chemical reference parameters and bioindicators was analysed to characterize the wastewater quality and composition. Results of the RT-qPCR based gene analysis indicate the presence of SARS-CoV-2 genetic traces in different raw wastewaters. Furthermore, selected samples have been sequenced using Sanger technology to confirm the specificity of the RT-qPCR and the origin of the coronavirus. A comparison of the particle-bound and the dissolved portion of SARS-CoV-2 virus genes shows that quantifications must not neglect the solid-phase reservoir. The infectivity of the raw wastewater has also been assessed by viral outgrowth assay with a potential SARS-CoV-2 host cell line in vitro, which were not infected when exposed to the samples. This first evidence suggests that wastewater might be no major route for transmission to humans. Our findings draw attention to the need for further methodological and molecular assay validation for enveloped viruses in wastewater.
Keywords: COVID-19; SARS-CoV-2; SARS-CoV-2 replication in vitro; Wastewater treatment; Wastewater-based epidemiology (WBE).
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Bar Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Paitan Y., Bitkover E., Berchenko Y., Kushmaro A. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.1101/2020.04.26.20073569. (2020.04.26.20073569) - DOI - PMC - PubMed
-
- Brückner I., Kirchner K., Müller Y., Schiwy S., Klaer K., Dolny R., Wendt L., Könemann S., Pinnekamp J., Hollert H. Status quo report on wastewater treatment plant, receiving water’s biocoenosis and quality as basis for evaluation of large-scale ozonation process. Water Sci. Technol. 2018;77(2):337–345. doi: 10.2166/wst.2017.548. - DOI - PubMed
-
- Carducci A., Federigi I., Liu D., Thompson J.R., Verani M. Making Waves: coronavirus detection, presence and persistence in the water environment: state of the art and knowledge needs for public health. Water Res. 2020;179:115907. doi: 10.1016/j.watres.2020.115907. 15 July 2020. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

