Small-molecule inducible transcriptional control in mammalian cells
- PMID: 32862714
- PMCID: PMC7914288
- DOI: 10.1080/07388551.2020.1808583
Small-molecule inducible transcriptional control in mammalian cells
Abstract
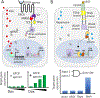
Tools for tuning transcription in mammalian cells have broad applications, from basic biological discovery to human gene therapy. While precise control over target gene transcription via dosing with small molecules (drugs) is highly sought, the design of such inducible systems that meets required performance metrics poses a great challenge in mammalian cell synthetic biology. Important characteristics include tight and tunable gene expression with a low background, minimal drug toxicity, and orthogonality. Here, we review small-molecule-inducible transcriptional control devices that have demonstrated success in mammalian cells and mouse models. Most of these systems employ natural or designed ligand-binding protein domains to directly or indirectly communicate with transcription machinery at a target sequence, via carefully constructed fusions. Example fusions include those to transcription activator-like effectors (TALEs), DNA-targeting proteins (e.g. dCas systems) fused to transactivating domains, and recombinases. Similar to the architecture of Type I nuclear receptors, many of the systems are designed such that the transcriptional controller is excluded from the nucleus in the absence of an inducer. Techniques that use ligand-induced proteolysis and antibody-based chemically induced dimerizers are also described. Collectively, these transcriptional control devices take advantage of a variety of recently developed molecular biology tools and cell biology insights and represent both proof of concept (e.g. targeting reporter gene expression) and disease-targeting studies.
Keywords: CAR T cells; TetR; CRISPRa/CRISPRi; TALEs; bacterial transcriptional factors; hormone receptors.
Figures

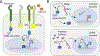
Similar articles
-
Small molecule-inducible gene regulatory systems in mammalian cells: progress and design principles.Curr Opin Biotechnol. 2022 Dec;78:102823. doi: 10.1016/j.copbio.2022.102823. Epub 2022 Oct 27. Curr Opin Biotechnol. 2022. PMID: 36332343 Free PMC article. Review.
-
Design and in vitro characterization of a single regulatory module for efficient control of gene expression in both plasmid DNA and a self-inactivating lentiviral vector.Mol Med. 2001 Aug;7(8):569-79. Mol Med. 2001. PMID: 11591893 Free PMC article.
-
Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors.ACS Synth Biol. 2014 Oct 17;3(10):723-30. doi: 10.1021/sb400114p. Epub 2013 Nov 22. ACS Synth Biol. 2014. PMID: 24251925 Free PMC article.
-
Amplification of small molecule-inducible gene expression via tuning of intracellular receptor densities.Nucleic Acids Res. 2015 Feb 18;43(3):1955-64. doi: 10.1093/nar/gku1388. Nucleic Acids Res. 2015. PMID: 25589545 Free PMC article.
-
TALE: a tale of genome editing.Prog Biophys Mol Biol. 2014 Jan;114(1):25-32. doi: 10.1016/j.pbiomolbio.2013.11.006. Epub 2013 Nov 27. Prog Biophys Mol Biol. 2014. PMID: 24291598 Review.
Cited by
-
All-in-one IQ toggle switches with high versatilities for fine-tuning of transgene expression in mammalian cells and tissues.Mol Ther Methods Clin Dev. 2024 Feb 2;32(1):101202. doi: 10.1016/j.omtm.2024.101202. eCollection 2024 Mar 14. Mol Ther Methods Clin Dev. 2024. PMID: 38374964 Free PMC article.
-
Small molecule-inducible gene regulatory systems in mammalian cells: progress and design principles.Curr Opin Biotechnol. 2022 Dec;78:102823. doi: 10.1016/j.copbio.2022.102823. Epub 2022 Oct 27. Curr Opin Biotechnol. 2022. PMID: 36332343 Free PMC article. Review.
-
Engineering autonomous closed-loop designer cells for disease therapy.iScience. 2022 Jan 29;25(3):103834. doi: 10.1016/j.isci.2022.103834. eCollection 2022 Mar 18. iScience. 2022. PMID: 35243222 Free PMC article. Review.
-
IQ-Switch is a QF-based innocuous, silencing-free, and inducible gene switch system in zebrafish.Commun Biol. 2021 Dec 16;4(1):1405. doi: 10.1038/s42003-021-02923-3. Commun Biol. 2021. PMID: 34916605 Free PMC article.
-
Engineering T Cell Development for the Next Generation of Stem Cell-Derived Immunotherapies.GEN Biotechnol. 2023 Apr 1;2(2):106-119. doi: 10.1089/genbio.2023.0008. Epub 2023 Apr 18. GEN Biotechnol. 2023. PMID: 37928777 Free PMC article. Review.
References
-
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, et al., Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993, 73, 585–596. - PubMed
-
- Wu S, Huang J, Dong J, Pan D, hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
