Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p
- PMID: 32863766
- PMCID: PMC7448321
- DOI: 10.1186/s12935-020-01501-7
Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p
Abstract
Background: Oral squamous cell carcinoma (OSCC) is a more common cancer in the world. Emerging evidence suggests that circular RNAs (circRNAs) participate in the progression of OSCC. However, the role of circ_0000140 in OSCC is still unknown.
Methods: The expression of circ_0000140 and microRNA-182-5p (miR-182-5p) were assessed by quantitative real-time polymerase chain reaction (qRT-PCR). Also, cell proliferation, migration and invasion were measured by colony formation and transwell assays, respectively. Western blot (WB) analysis was used to test the levels of proliferation, metastasis and glycolysis metabolism-related proteins as well as cell division cycle 73 (CDC73) protein. Further, the extracellular acidification rate (ECAR) of cells was detected by the Seahorse XF Extracellular Flux Analyzer. The lactate acid level of cells was tested by Lactate Assay Kit. Moreover, dual-luciferase reporter was used to verify the interaction between miR-182-3p and circ_0000140 or CDC73, and RNA immunoprecipitation (RIP) assay was employed to further confirm the relationship between miR-182-3p and circ_0000140. In addition, mice xenograft models were built to measure the effect of circ_0000140 on OSCC tumor growth in vivo.
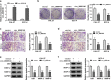
Results: Circ_0000140 was lowly expressed in OSCC, and its overexpression hindered proliferation, migration, invasion and glycolysis metabolism in OSCC cells. MiR-182-5p could be sponged by circ_0000140, and its mimic could invert the suppression of circ_0000140 overexpression on OSCC progression. CDC73 could be targeted by miR-182-3p, and its silencing could reverse the inhibition of miR-182-3p inhibitor on OSCC progression. Further, overexpressed circ_0000140 reduced the OSCC tumor growth in vivo.
Conclusions: Circ_0000140 might play an anti-cancer role in OSCC, which provided a novel target for clinical therapy of OSCC.
Keywords: CDC73; OSCC; circ_0000140; miR-182-5p.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures







References
-
- Dos Reis PP, Bharadwaj RR, Machado J, Macmillan C, Pintilie M, Sukhai MA, Perez-Ordonez B, Gullane P, Irish J, Kamel-Reid S. Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer. 2008;113(11):3169–80. doi: 10.1002/cncr.23934. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

