Comprehensive analysis of a long non-coding RNA-associated competing endogenous RNA network in glioma
- PMID: 32863896
- PMCID: PMC7436175
- DOI: 10.3892/ol.2020.11924
Comprehensive analysis of a long non-coding RNA-associated competing endogenous RNA network in glioma
Abstract
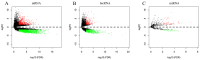
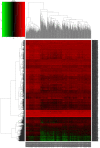
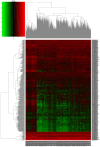
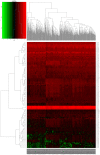
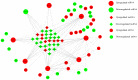
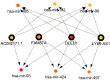
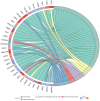
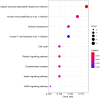
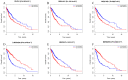
Long non-coding RNAs (lncRNAs) can act as competing endogenous RNAs (ceRNAs), interacting with microRNAs (miRNAs) and playing an important role in tumor progression. However, the role of lncRNA-mediated ceRNAs in glioma remains largely unknown. The present study aimed to identify novel lncRNAs and their associated function in glioma. RNA sequencing and corresponding clinical data from patients with glioma were obtained from The Cancer Genome Atlas. A total of 598 glioma tissues and 5 normal brain tissues were analyzed in the present study. The differentially expressed (DE) lncRNAs, mRNAs and miRNAs were identified using R packages and were used to construct a ceRNA network. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed to investigate the biological functions of the DEmRNAs. Kaplan-Meier curve analysis was performed to investigate the association between DElncRNA expression and patient outcome. A total of 752 DElncRNAs, 2,079 DEmRNAs and 113 DEmiRNAs were identified between glioma and normal tissues. A lncRNA-miRNA-mRNA ceRNA network consisting of 61 lncRNAs, 12 miRNAs and 92 mRNAs was constructed. Survival analysis indicated that 36 DElncRNAs, 72 DEmRNAs and 3 DEmiRNAs were associated with overall survival in patients with glioma. The present study identified novel lncRNAs associated with survival prognosis and may facilitate further investigation of lncRNA-mediated ceRNA regulatory mechanisms in glioma.
Keywords: competitive endogenous RNA; glioma; long non-coding RNA; survival analysis.
Copyright: © Zhu et al.
Figures
References
-
- National Brain Tumor Association, corp-author. Quick brain tumor facts. [Nov 27;2017 ];2018 Available at: http: //braintumor.org/brain-tumor-information/brain-tumor-facts/
-
- American Brain Tumor Association (ABTA), corp-author Brain tumor statistics. [Nov 22;2017 ];2017 Available at: http://www.abta.org/about-us/news/brain-tumor-statistics/
LinkOut - more resources
Full Text Sources
