Jupyter notebook-based tools for building structured datasets from the Sequence Read Archive
- PMID: 32864105
- PMCID: PMC7445559
- DOI: 10.12688/f1000research.23180.2
Jupyter notebook-based tools for building structured datasets from the Sequence Read Archive
Abstract
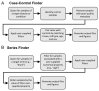
The Sequence Read Archive (SRA) is a large public repository that stores raw next-generation sequencing data from thousands of diverse scientific investigations. Despite its promise, reuse and re-analysis of SRA data has been challenged by the heterogeneity and poor quality of the metadata that describe its biological samples. Recently, the MetaSRA project standardized these metadata by annotating each sample with terms from biomedical ontologies. In this work, we present a pair of Jupyter notebook-based tools that utilize the MetaSRA for building structured datasets from the SRA in order to facilitate secondary analyses of the SRA's human RNA-seq data. The first tool, called the Case-Control Finder, finds suitable case and control samples for a given disease or condition where the cases and controls are matched by tissue or cell type. The second tool, called the Series Finder, finds ordered sets of samples for the purpose of addressing biological questions pertaining to changes over a numerical property such as time. These tools were the result of a three-day-long NCBI Codeathon in March 2019 held at the University of North Carolina at Chapel Hill.
Keywords: Hackathon; Jupyter; MetaSRA; Metadata; Ontology; RNA-seq; Sequence Read Archive.
Copyright: © 2020 Bernstein MN et al.
Conflict of interest statement
No competing interests were disclosed.
Figures
Similar articles
-
MetaSRA: normalized human sample-specific metadata for the Sequence Read Archive.Bioinformatics. 2017 Sep 15;33(18):2914-2923. doi: 10.1093/bioinformatics/btx334. Bioinformatics. 2017. PMID: 28535296 Free PMC article.
-
pysradb: A Python package to query next-generation sequencing metadata and data from NCBI Sequence Read Archive.F1000Res. 2019 Apr 23;8:532. doi: 10.12688/f1000research.18676.1. eCollection 2019. F1000Res. 2019. PMID: 31114675 Free PMC article.
-
The CAIRR Pipeline for Submitting Standards-Compliant B and T Cell Receptor Repertoire Sequencing Studies to the National Center for Biotechnology Information Repositories.Front Immunol. 2018 Aug 16;9:1877. doi: 10.3389/fimmu.2018.01877. eCollection 2018. Front Immunol. 2018. PMID: 30166985 Free PMC article.
-
Tools for the analysis of high-dimensional single-cell RNA sequencing data.Nat Rev Nephrol. 2020 Jul;16(7):408-421. doi: 10.1038/s41581-020-0262-0. Epub 2020 Mar 27. Nat Rev Nephrol. 2020. PMID: 32221477 Review.
-
The eXtensible ontology development (XOD) principles and tool implementation to support ontology interoperability.J Biomed Semantics. 2018 Jan 12;9(1):3. doi: 10.1186/s13326-017-0169-2. J Biomed Semantics. 2018. PMID: 29329592 Free PMC article. Review.
Cited by
-
Microbial Dark Matter: from Discovery to Applications.Genomics Proteomics Bioinformatics. 2022 Oct;20(5):867-881. doi: 10.1016/j.gpb.2022.02.007. Epub 2022 Apr 26. Genomics Proteomics Bioinformatics. 2022. PMID: 35477055 Free PMC article. Review.
-
Enhancing Learning About Epidemiological Data Analysis Using R for Graduate Students in Medical Fields With Jupyter Notebook: Classroom Action Research.JMIR Med Educ. 2023 May 29;9:e47394. doi: 10.2196/47394. JMIR Med Educ. 2023. PMID: 37247206 Free PMC article.
-
Detection and Classification of Melanoma Skin Cancer Using Image Processing Technique.Diagnostics (Basel). 2023 Oct 26;13(21):3313. doi: 10.3390/diagnostics13213313. Diagnostics (Basel). 2023. PMID: 37958209 Free PMC article.
-
Metadata retrieval from sequence databases with ffq.Bioinformatics. 2023 Jan 1;39(1):btac667. doi: 10.1093/bioinformatics/btac667. Bioinformatics. 2023. PMID: 36610997 Free PMC article.
-
STAT: a fast, scalable, MinHash-based k-mer tool to assess Sequence Read Archive next-generation sequence submissions.Genome Biol. 2021 Sep 20;22(1):270. doi: 10.1186/s13059-021-02490-0. Genome Biol. 2021. PMID: 34544477 Free PMC article.
References
-
- Bernstein M: mbernste/hypothesis-driven-SRA-queries: First release (Version v1.0.0). Zenodo. 2020. 10.5281/zenodo.3807512 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

