Dynamic full-field optical coherence tomography: 3D live-imaging of retinal organoids
- PMID: 32864115
- PMCID: PMC7429964
- DOI: 10.1038/s41377-020-00375-8
Dynamic full-field optical coherence tomography: 3D live-imaging of retinal organoids
Abstract
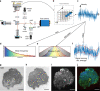
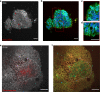
Optical coherence tomography offers astounding opportunities to image the complex structure of living tissue but lacks functional information. We present dynamic full-field optical coherence tomography as a technique to noninvasively image living human induced pluripotent stem cell-derived retinal organoids. Coloured images with an endogenous contrast linked to organelle motility are generated, with submicrometre spatial resolution and millisecond temporal resolution, creating a way to identify specific cell types in living tissue via their function.
Keywords: Imaging and sensing; Interference microscopy; Wide-field fluorescence microscopy.
© The Author(s) 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

References
-
- Hajdu SI. The first use of the microscope in medicine. Ann. Clin. Lab. Sci. 2002;32:309–310. - PubMed
-
- Jedrzejczak-Silicka, M. History of cell culture in New Insights into Cell Culture Technology. ed. Sivakumar Joghi Thatha Gowder, IntechOpen, (2017).
LinkOut - more resources
Full Text Sources

