Aberrant RNA Splicing in Cancer
- PMID: 32864546
- PMCID: PMC7453310
- DOI: 10.1146/annurev-cancerbio-030617-050407
Aberrant RNA Splicing in Cancer
Abstract
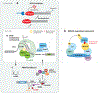
RNA splicing, the enzymatic process of removing segments of premature RNA to produce mature RNA, is a key mediator of proteome diversity and regulator of gene expression. Increased systematic sequencing of the genome and transcriptome of cancers has identified a variety of means by which RNA splicing is altered in cancer relative to normal cells. These findings, in combination with the discovery of recurrent change-of-function mutations in splicing factors in a variety of cancers, suggest that alterations in splicing are drivers of tumorigenesis. Greater characterization of altered splicing in cancer parallels increasing efforts to pharmacologically perturb splicing and early-phase clinical development of small molecules that disrupt splicing in patients with cancer. Here we review recent studies of global changes in splicing in cancer, splicing regulation of mitogenic pathways critical in cancer transformation, and efforts to therapeutically target splicing in cancer.
Keywords: RNA; SF3B1; SRSF2; U2AF1; ZRSR2; splicing.
Figures




References
-
- Agrawal AA, Yu L, Smith PG, Buonamici S. 2018. Targeting splicing abnormalities in cancer. Curr. Opin. Genet. Dev 48:67–74 - PubMed
-
- Agrawal S, Eng C. 2006. Differential expression of novel naturally occurring splice variants of PTEN and their functional consequences in Cowden syndrome and sporadic breast cancer. um. Mol. Genet 15:777–87 - PubMed
-
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, et al. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391–405 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
