Discovery and Verification of Extracellular miRNA Biomarkers for Non-invasive Prediction of Pre-eclampsia in Asymptomatic Women
- PMID: 32864636
- PMCID: PMC7455024
- DOI: 10.1016/j.xcrm.2020.100013
Discovery and Verification of Extracellular miRNA Biomarkers for Non-invasive Prediction of Pre-eclampsia in Asymptomatic Women
Abstract
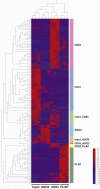
Development of effective prevention and treatment strategies for pre-eclampsia is limited by the lack of accurate methods for identification of at-risk pregnancies. We performed small RNA sequencing (RNA-seq) of maternal serum extracellular RNAs (exRNAs) to discover and verify microRNAs (miRNAs) differentially expressed in patients who later developed pre-eclampsia. Sera collected from 73 pre-eclampsia cases and 139 controls between 17 and 28 weeks gestational age (GA), divided into separate discovery and verification cohorts, are analyzed by small RNA-seq. Discovery and verification of univariate and bivariate miRNA biomarkers reveal that bivariate biomarkers verify at a markedly higher rate than univariate biomarkers. The majority of verified biomarkers contain miR-155-5p, which has been reported to mediate the pre-eclampsia-associated repression of endothelial nitric oxide synthase (eNOS) by tumor necrosis factor alpha (TNF-α). Deconvolution analysis reveals that several verified miRNA biomarkers come from the placenta and are likely carried by placenta-specific extracellular vesicles.
Conflict of interest statement
L.C.L. is a site PI for the Sera Prognostics TREETOP study. J.B.B., J.B., D.E.H., A.C.F., M.T.D., S.R.R., A.J.L., and R.T. are employees and shareholders in Sera Prognostics. Sera Prognostics has filed a provisional patent application (United States Provisional Application no. 62/777,576) on the findings presented in this manuscript.
Figures



References
-
- Ness R.B., Roberts J.M. Heterogeneous causes constituting the single syndrome of preeclampsia: a hypothesis and its implications. Am. J. Obstet. Gynecol. 1996;175:1365–1370. - PubMed
-
- Sibai B.M. Diagnosis and management of gestational hypertension and preeclampsia. Obstet. Gynecol. 2003;102:181–192. - PubMed
-
- Sibai B.M. Preeclampsia as a cause of preterm and late preterm (near-term) births. Semin. Perinatol. 2006;30:16–19. - PubMed
-
- Henderson J.T., Whitlock E.P., O’Conner E., Senger C.A., Thompson J.H., Rowland M.G. Low-dose aspirin for prevention of morbidity and mortality from preeclampsia: a systematic evidence review for the U.S. Preventive Services Task Force. Ann. Intern. Med. 2014;160:695–703. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

